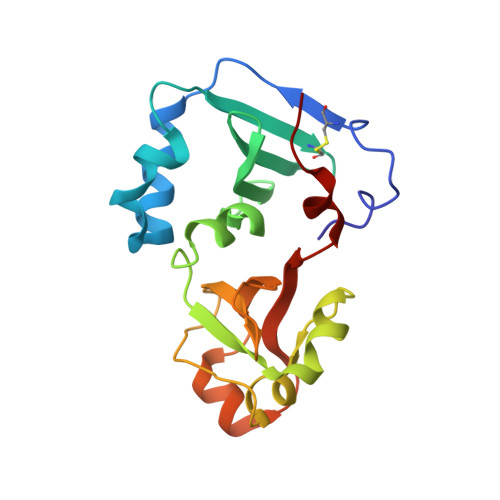
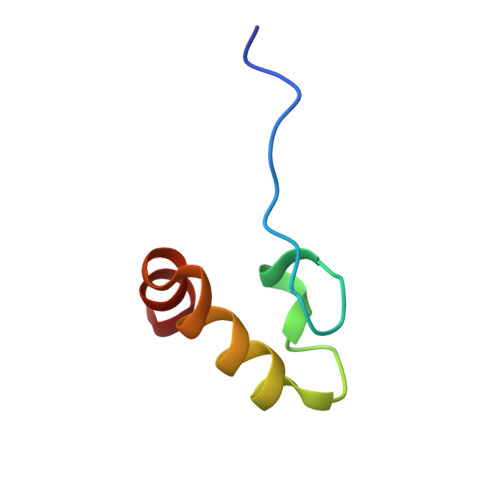
Crystal structure of the DENR-MCT-1 complex revealed zinc-binding site essential for heterodimer formation.
Lomakin, I.B., Dmitriev, S.E., Steitz, T.A.(2019) Proc Natl Acad Sci U S A 116: 528-533
- PubMed: 30584092
- DOI: https://doi.org/10.1073/pnas.1809688116
- Primary Citation of Related Structures:
6MS4 - PubMed Abstract:
The density-regulated protein (DENR) and the malignant T cell-amplified sequence 1 (MCT-1/MCTS1) oncoprotein support noncanonical translation initiation, promote translation reinitiation on a specific set of mRNAs with short upstream reading frames, and regulate ribosome recycling. DENR and MCT-1 form a heterodimer, which binds to the ribosome. We determined the crystal structure of the heterodimer formed by human MCT-1 and the N-terminal domain of DENR at 2.0-Å resolution. The structure of the heterodimer reveals atomic details of the mechanism of DENR and MCT-1 interaction. Four conserved cysteine residues of DENR (C34, C37, C44, C53) form a classical tetrahedral zinc ion-binding site, which preserves the structure of the DENR's MCT-1-binding interface that is essential for the dimerization. Substitution of all four cysteines by alanine abolished a heterodimer formation. Our findings elucidate further the mechanism of regulation of DENR-MCT-1 activities in unconventional translation initiation, reinitiation, and recycling.
Organizational Affiliation:
Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520-8114; ivan.lomakin@yale.edu.