CH7233163 Overcomes Osimertinib-Resistant EGFR-Del19/T790M/C797S Mutation.
Kashima, K., Kawauchi, H., Tanimura, H., Tachibana, Y., Chiba, T., Torizawa, T., Sakamoto, H.(2020) Mol Cancer Ther 19: 2288-2297
- PubMed: 32943545
- DOI: https://doi.org/10.1158/1535-7163.MCT-20-0229
- Primary Citation of Related Structures:
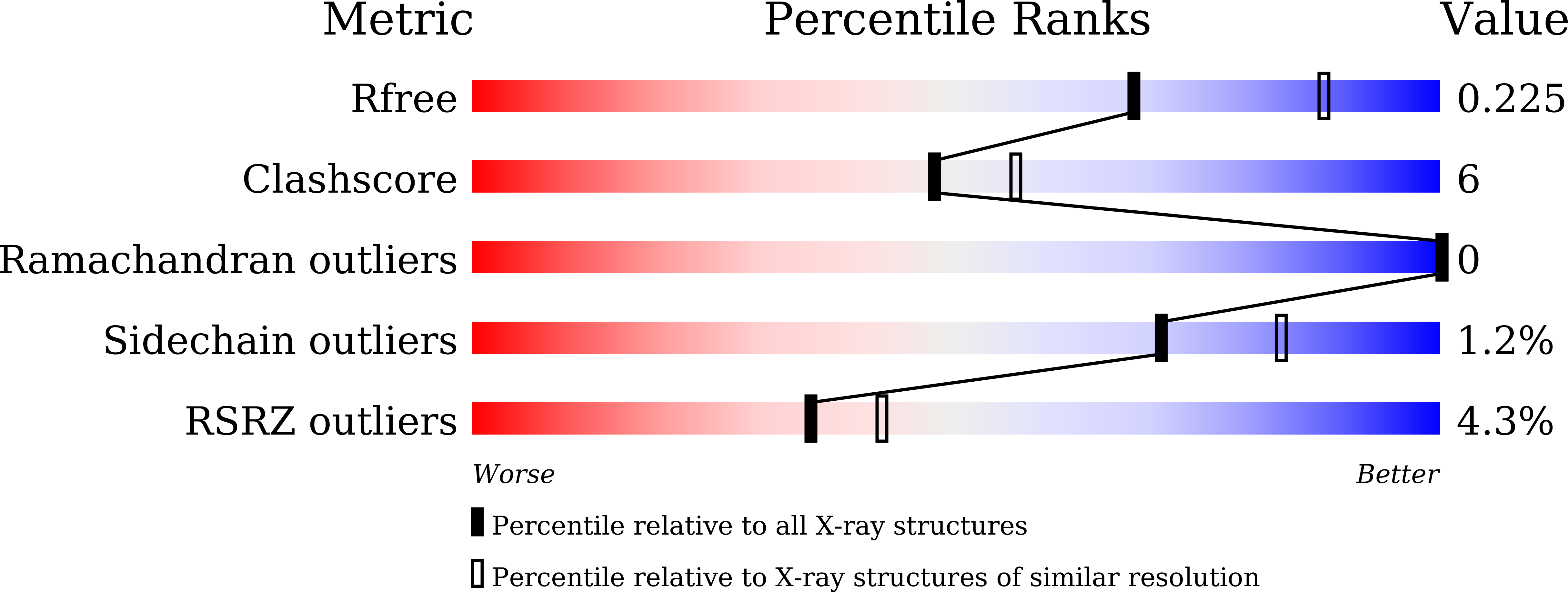
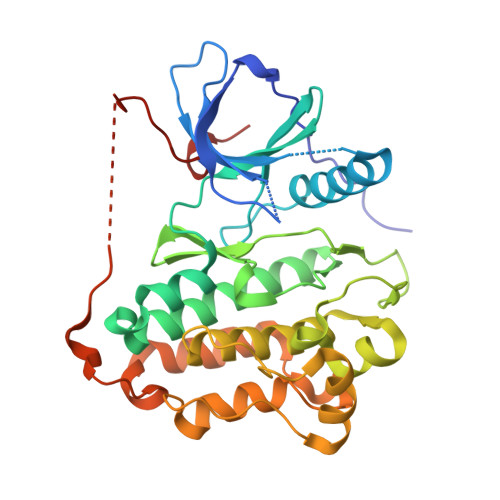
6LUB, 6LUD - PubMed Abstract:
Osimertinib is the only EGFR-tyrosine kinase inhibitor (TKI) capable of overcoming EGFR-T790M-mutated NSCLC, but osimertinib-resistant EGFR triple mutations (Del19/T790M/C797S or L858R/T790M/C797S) have been reported. Although allosteric EGFR TKIs (e.g., EAI-045) that potentially overcome L858R/T790M/C797S have been identified, there are no effective inhibitors against Del19/T790M/C797S. In this study, we identified CH7233163 as having the potential to overcome EGFR-Del19/T790M/C797S. CH7233163 showed potent antitumor activities against tumor with EGFR-Del19/T790M/C797S in vitro and in vivo In addition to EGFR-Del19/T790M/C797S, the characterization assays showed that CH7233163 more selectively inhibits various types of EGFR mutants (e.g., L858R/T790M/C797S, L858R/T790M, Del19/T790M, Del19, and L858R) over wild type. Furthermore, crystal structure analysis suggested that CH7233163 is a noncovalent ATP-competitive inhibitor for EGFR-Del19/T790M/C797S that utilizes multiple interactions with the EGFR's αC-helix-in conformation to achieve potent inhibitory activity and mutant selectivity. Therefore, we conclude that CH7233163 is a potentially effective therapy for osimertinib-resistant patients, especially in cases of EGFR-Del19/T790M/C797S.
Organizational Affiliation:
Research Division, Chugai Pharmaceutical Co. Ltd., Kanagawa, Japan. kashimaknj@chugai-pharm.co.jp.