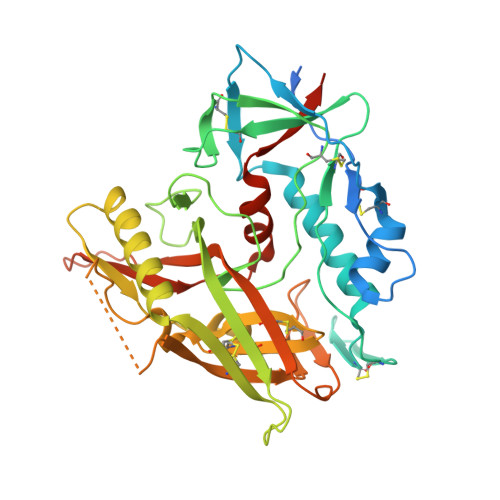
A non-canonical binding interface in the crystal structure of HIV-1 gp120 core in complex with CD4.
Duan, L.W., Zhang, H., Zhao, M.T., Sun, J.X., Chen, W.L., Lin, J.P., Liu, X.Q.(2017) Sci Rep 7: 46733-46733
- PubMed: 28429756
- DOI: https://doi.org/10.1038/srep46733
- Primary Citation of Related Structures:
6L1Y - PubMed Abstract:
Numerous crystal structures of HIV gp120 have been reported, alone or with receptor CD4 and cognate antibodies; however, no sole gp120/CD4 complex without stabilization by an antibody is available. Here, we report a crystal structure of the gp120/CD4 complex without the aid of an antibody from HIV-1 CRF07_BC, a strain circulating in China. Interestingly, in addition to the canonical binding surface, a second interacting interface was identified. A mutagenesis study on critical residues revealed that the stability of this interface is important for the efficiency of Env-mediated membrane fusion. Furthermore, we found that a broad neutralizing antibody, ibalizumab, which targets CD4 in the absence of gp120, occupies the same binding surface as the second interface identified here on gp120. Therefore, we identified the possibility of the involvement of a second gp120-CD4 interaction interface during viral entry, and also provided a reasonable explanation for the broad activity of neutralizing antibody ibalizumab.
Organizational Affiliation:
State Key Laboratory of Medicinal Chemical Biology, College of Life Sciences, Nankai University, Tianjin 300071, China.