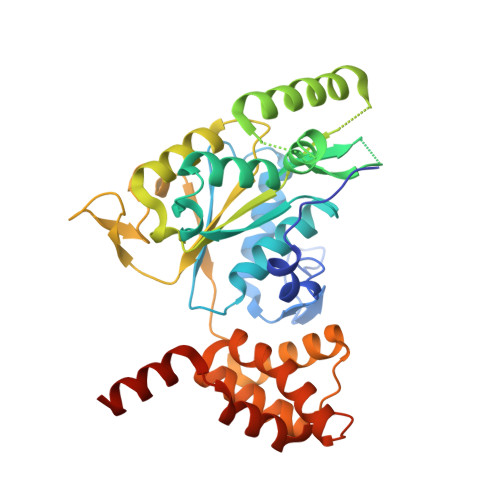
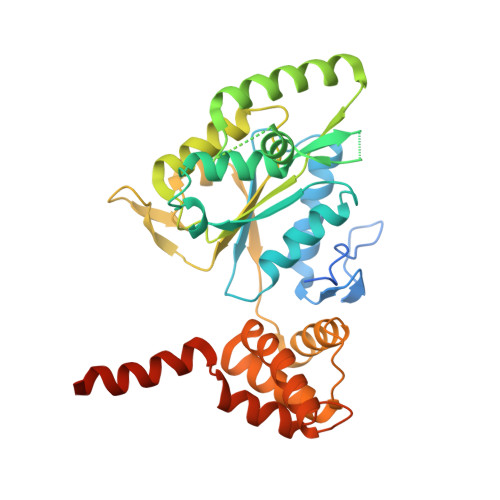
Chemical perturbations reveal that RUVBL2 regulates the circadian phase in mammals.
Ju, D., Zhang, W., Yan, J., Zhao, H., Li, W., Wang, J., Liao, M., Xu, Z., Wang, Z., Zhou, G., Mei, L., Hou, N., Ying, S., Cai, T., Chen, S., Xie, X., Lai, L., Tang, C., Park, N., Takahashi, J.S., Huang, N., Qi, X., Zhang, E.E.(2020) Sci Transl Med 12
- PubMed: 32376767
- DOI: https://doi.org/10.1126/scitranslmed.aba0769
- Primary Citation of Related Structures:
6K0R - PubMed Abstract:
Transcriptional regulation lies at the core of the circadian clockwork, but how the clock-related transcription machinery controls the circadian phase is not understood. Here, we show both in human cells and in mice that RuvB-like ATPase 2 (RUVBL2) interacts with other known clock proteins on chromatin to regulate the circadian phase. Pharmacological perturbation of RUVBL2 with the adenosine analog compound cordycepin resulted in a rapid-onset 12-hour clock phase-shift phenotype at human cell, mouse tissue, and whole-animal live imaging levels. Using simple peripheral injection treatment, we found that cordycepin penetrated the blood-brain barrier and caused rapid entrainment of the circadian phase, facilitating reduced duration of recovery in a mouse jet-lag model. We solved a crystal structure for human RUVBL2 in complex with a physiological metabolite of cordycepin, and biochemical assays showed that cordycepin treatment caused disassembly of an interaction between RUVBL2 and the core clock component BMAL1. Moreover, we showed with spike-in ChIP-seq analysis and binding assays that cordycepin treatment caused disassembly of the circadian super-complex, which normally resides at E-box chromatin loci such as PER1 , PER2 , DBP, and NR1D1 Mathematical modeling supported that the observed type 0 phase shifts resulted from derepression of E-box clock gene transcription.
Organizational Affiliation:
National Institute of Biological Sciences, Beijing 102206, China.