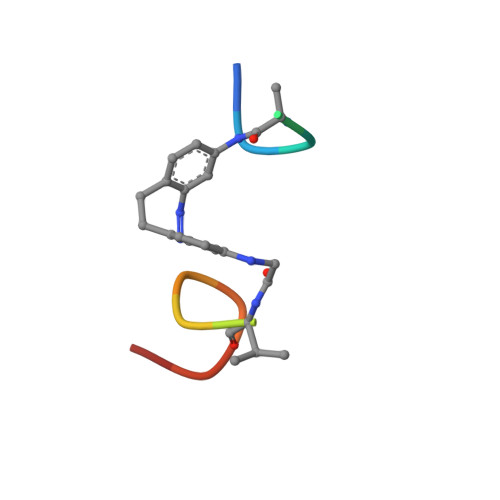
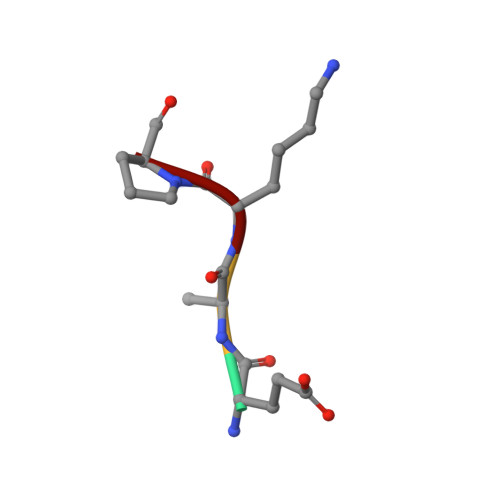
Modulating Protein-Protein Interactions with Visible-Light-Responsive Peptide Backbone Photoswitches.
Albert, L., Penalver, A., Djokovic, N., Werel, L., Hoffarth, M., Ruzic, D., Xu, J., Essen, L.O., Nikolic, K., Dou, Y., Vazquez, O.(2019) Chembiochem 20: 1417-1429
- PubMed: 30675988
- DOI: https://doi.org/10.1002/cbic.201800737
- Primary Citation of Related Structures:
6IAM - PubMed Abstract:
Life relies on a myriad of carefully orchestrated processes, in which proteins and their direct interplay ultimately determine cellular function and disease. Modulation of this complex crosstalk has recently attracted attention, even as a novel therapeutic strategy. Herein, we describe the synthesis and characterization of two visible-light-responsive peptide backbone photoswitches based on azobenzene derivatives, to exert optical control over protein-protein interactions (PPI). The novel peptidomimetics undergo fast and reversible isomerization with low photochemical fatigue under alternatively blue-/green-light irradiation cycles. Both bind in the nanomolar range to the protein of interest. Importantly, the best peptidomimetic displays a clear difference between isomers in its protein-binding capacity and, in turn, in its potential to inhibit enzymatic activity through PPI disruption. In addition, crystal structure determination, docking and molecular dynamics calculations allow a molecular interpretation and open up new avenues in the design and synthesis of future photoswitchable PPI modulators.
Organizational Affiliation:
Fachbereich Chemie, Philipps-Universität Marburg, Hans-Meerwein-Strasse 4, 35043, Marburg, Germany.