Crystal structure of the WT human mitochondrial football Hsp60-Hsp10(ADPBeFx)14 complex
Jebara, F., Patra, M., Azem, A.(2020) Nat Commun
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2020) Nat Commun
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60 kDa heat shock protein, mitochondrial | 549 | Homo sapiens | Mutation(s): 0 Gene Names: HSPD1, HSP60 EC: 3.6.4.9 | 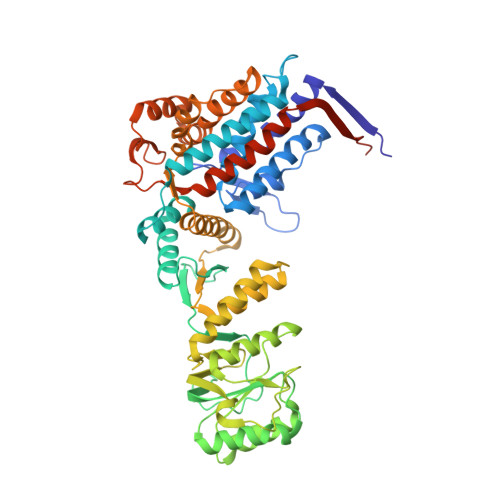 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P10809 (Homo sapiens) Explore P10809 Go to UniProtKB: P10809 | |||||
PHAROS: P10809 GTEx: ENSG00000144381 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P10809 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 10 kDa heat shock protein, mitochondrial | 102 | Homo sapiens | Mutation(s): 0 Gene Names: HSPE1 | 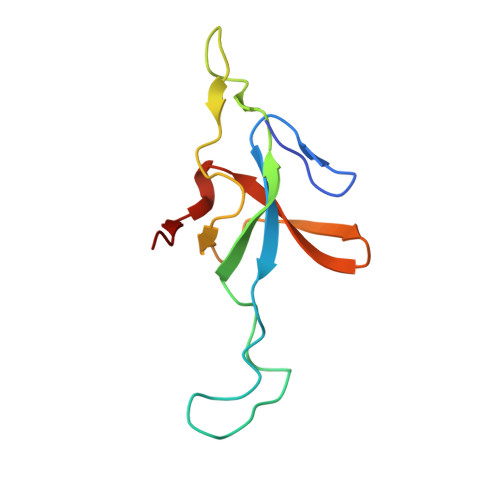 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P61604 (Homo sapiens) Explore P61604 Go to UniProtKB: P61604 | |||||
PHAROS: P61604 GTEx: ENSG00000115541 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P61604 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ADP Query on ADP | AB [auth K] CA [auth J] CC [auth A] EB [auth G] GA [auth I] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| BEF Query on BEF | BB [auth K] DA [auth J] DC [auth A] FB [auth G] HA [auth I] | BERYLLIUM TRIFLUORIDE ION Be F3 OGIAHMCCNXDTIE-UHFFFAOYSA-K |  | ||
| K Query on K | BC [auth B] DB [auth K] FA [auth J] FC [auth A] HB [auth G] | POTASSIUM ION K NPYPAHLBTDXSSS-UHFFFAOYSA-N |  | ||
| MG Query on MG | AC [auth B] CB [auth K] EA [auth J] EC [auth A] GB [auth G] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 141.59 | α = 90 |
| b = 295.78 | β = 90 |
| c = 326.53 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| PHASER | phasing |
| MxCuBE | data collection |
| Coot | model building |
| Funding Organization | Location | Grant Number |
|---|---|---|
| United States - Israel Binational Science Foundation (BSF) | Israel | 2015214 |