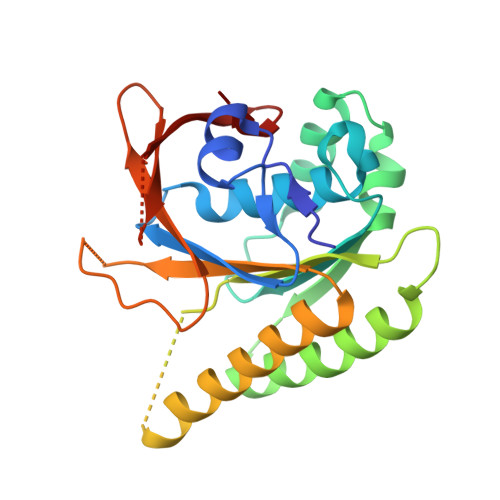
Improved RAD51 binders through motif shuffling based on the modularity of BRC repeats.
Lindenburg, L.H., Pantelejevs, T., Gielen, F., Zuazua-Villar, P., Butz, M., Rees, E., Kaminski, C.F., Downs, J.A., Hyvonen, M., Hollfelder, F.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34772801
- DOI: https://doi.org/10.1073/pnas.2017708118
- Primary Citation of Related Structures:
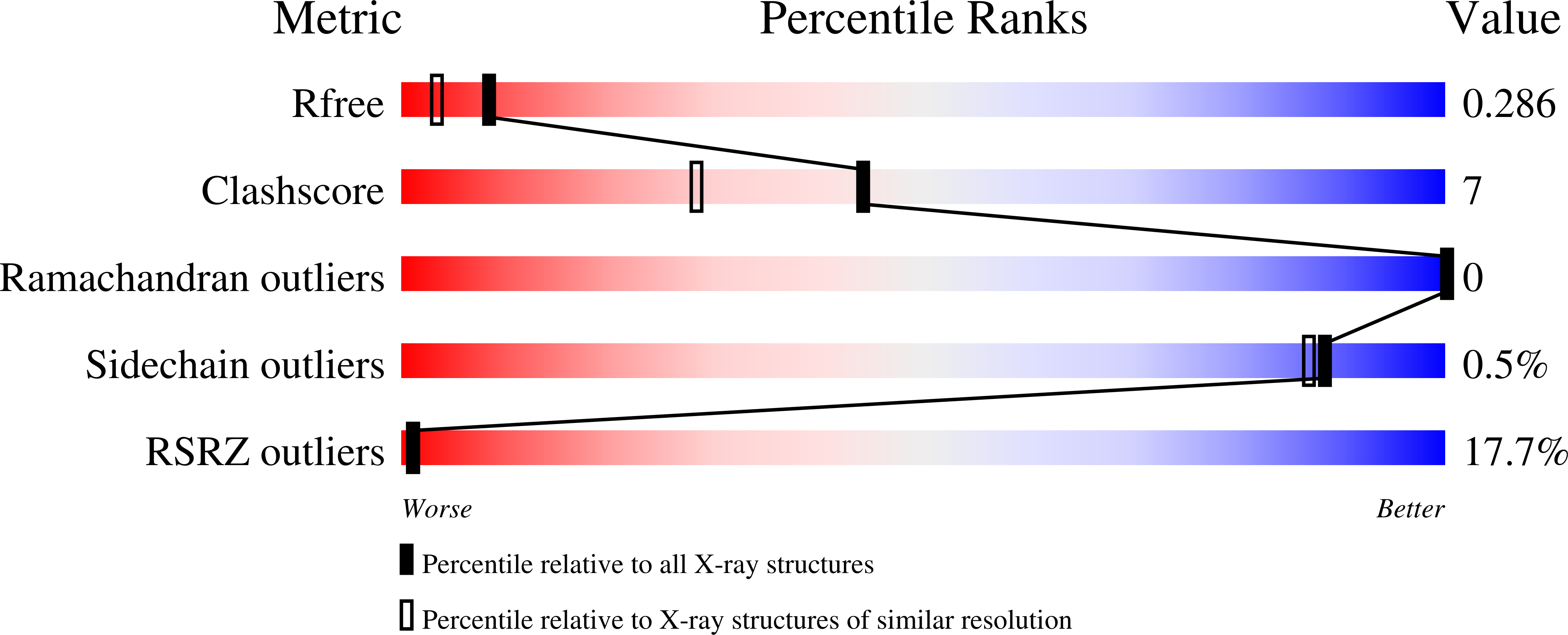
6HQU - PubMed Abstract:
Exchanges of protein sequence modules support leaps in function unavailable through point mutations during evolution. Here we study the role of the two RAD51-interacting modules within the eight binding BRC repeats of BRCA2. We created 64 chimeric repeats by shuffling these modules and measured their binding to RAD51. We found that certain shuffled module combinations were stronger binders than any of the module combinations in the natural repeats. Surprisingly, the contribution from the two modules was poorly correlated with affinities of natural repeats, with a weak BRC8 repeat containing the most effective N-terminal module. The binding of the strongest chimera, BRC8-2, to RAD51 was improved by -2.4 kCal/mol compared to the strongest natural repeat, BRC4. A crystal structure of RAD51:BRC8-2 complex shows an improved interface fit and an extended β-hairpin in this repeat. BRC8-2 was shown to function in human cells, preventing the formation of nuclear RAD51 foci after ionizing radiation.
Organizational Affiliation:
Department of Biochemistry, University of Cambridge, Cambridge CB2 1GA, United Kingdom.