3-Fluoro-4-hydroxyprolines: Synthesis, Conformational Analysis, and Stereoselective Recognition by the VHL E3 Ubiquitin Ligase for Targeted Protein Degradation.
Testa, A., Lucas, X., Castro, G.V., Chan, K.H., Wright, J.E., Runcie, A.C., Gadd, M.S., Harrison, W.T.A., Ko, E.J., Fletcher, D., Ciulli, A.(2018) J Am Chem Soc 140: 9299-9313
- PubMed: 29949369
- DOI: https://doi.org/10.1021/jacs.8b05807
- Primary Citation of Related Structures:
6GFX, 6GFY, 6GFZ - PubMed Abstract:
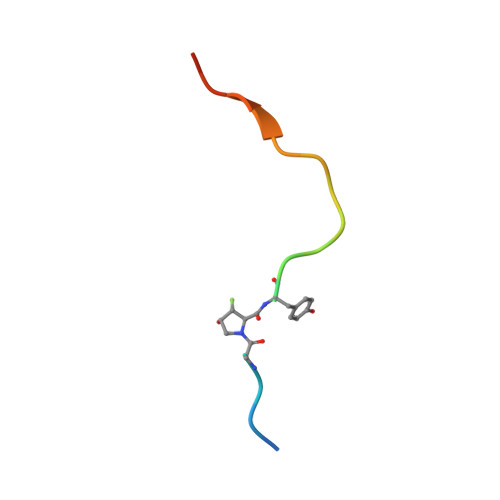
Hydroxylation and fluorination of proline alters the pyrrolidine ring pucker and the trans:cis amide bond ratio in a stereochemistry-dependent fashion, affecting molecular recognition of proline-containing molecules by biological systems. While hydroxyprolines and fluoroprolines are common motifs in medicinal and biological chemistry, the synthesis and molecular properties of prolines containing both modifications, i.e., fluoro-hydroxyprolines, have not been described. Here we present a practical and facile synthesis of all four diastereoisomers of 3-fluoro-4-hydroxyprolines (F-Hyps), starting from readily available 4-oxo-l-proline derivatives. Small-molecule X-ray crystallography, NMR spectroscopy, and quantum mechanical calculations are consistent with fluorination at C 3 having negligible effects on the hydrogen bond donor capacity of the C 4 hydroxyl, but inverting the natural preference of Hyp from C 4 -exo to C 4 -endo pucker. In spite of this, F-Hyps still bind to the von Hippel-Lindau (VHL) E3 ligase, which naturally recognizes C 4 -exo Hyp in a stereoselective fashion. Co-crystal structures and electrostatic potential calculations support and rationalize the observed preferential recognition for (3 R,4 S)-F-Hyp over the corresponding (3 S,4 S) epimer by VHL. We show that (3 R,4 S)-F-Hyp provides bioisosteric Hyp substitution in both hypoxia-inducible factor 1 alpha (HIF-1α) substrate peptides and peptidomimetic ligands that form part of PROTAC (proteolysis targeting chimera) conjugates for targeted protein degradation. Despite a weakened affinity, Hyp substitution with (3 S,4 S)-F-Hyp within the PROTAC MZ1 led to Brd4-selective cellular degradation at concentrations >100-fold lower than the binary K d for VHL. We anticipate that the disclosed chemistry of 3-fluoro-4-hydroxyprolines and their application as VHL ligands for targeted protein degradation will be of wide interest to medicinal organic chemists, chemical biologists, and drug discoverers alike.
Organizational Affiliation:
Division of Biological Chemistry and Drug Discovery, School of Life Sciences , University of Dundee , James Black Centre, Dow Street , Dundee DD1 5EH , Scotland, U.K.