Identification and Structure-Guided Development of Pyrimidinone Based USP7 Inhibitors.
O'Dowd, C.R., Helm, M.D., Rountree, J.S.S., Flasz, J.T., Arkoudis, E., Miel, H., Hewitt, P.R., Jordan, L., Barker, O., Hughes, C., Rozycka, E., Cassidy, E., McClelland, K., Odrzywol, E., Page, N., Feutren-Burton, S., Dvorkin, S., Gavory, G., Harrison, T.(2018) ACS Med Chem Lett 9: 238-243
- PubMed: 29541367
- DOI: https://doi.org/10.1021/acsmedchemlett.7b00512
- Primary Citation of Related Structures:
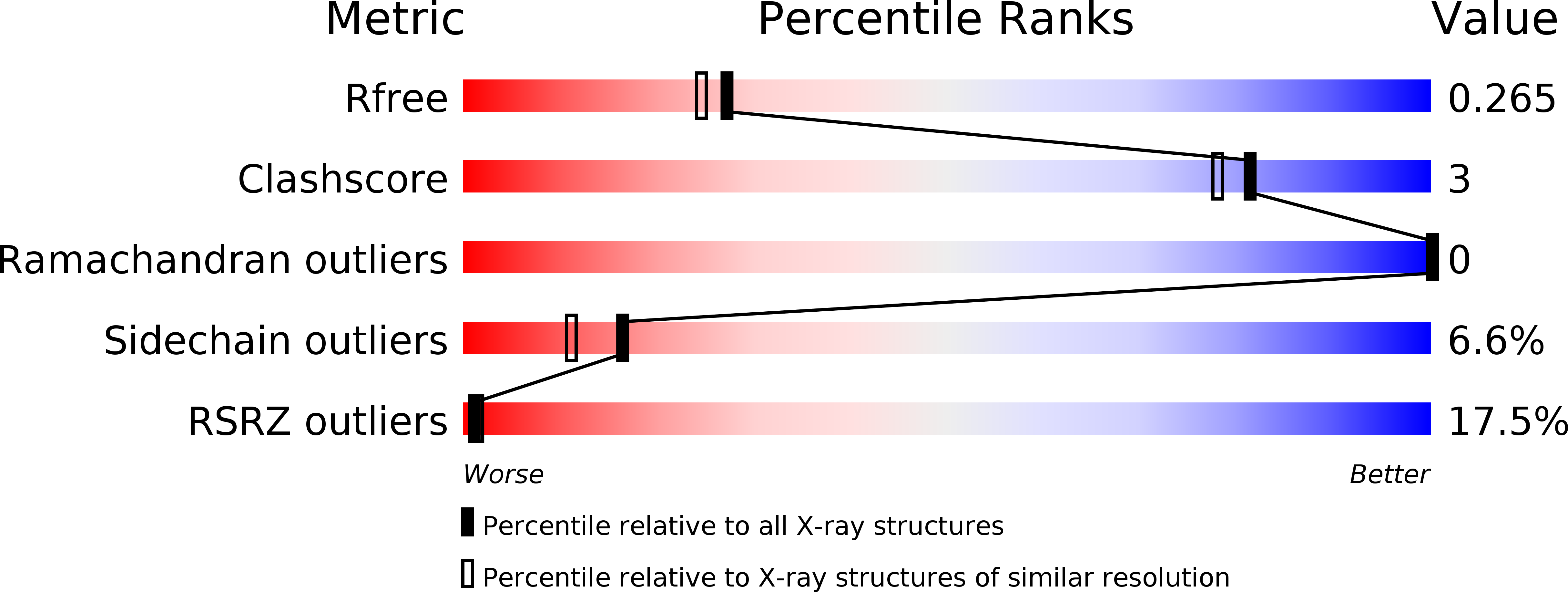
6F5H - PubMed Abstract:
Ubiquitin specific protease 7 (USP7, HAUSP) has become an attractive target in drug discovery due to the role it plays in modulating Mdm2 levels and consequently p53. Increasing interest in USP7 is emerging due to its potential involvement in oncogenic pathways as well as possible roles in both metabolic and immune disorders in addition to viral infections. Potent, novel, and selective inhibitors of USP7 have been developed using both rational and structure-guided design enabled by high-resolution cocrystallography. Initial hits were identified via fragment-based screening, scaffold-hopping, and hybridization exercises. Two distinct subseries are described along with associated structure-activity relationship trends, as are initial efforts aimed at developing compounds suitable for in vivo experiments. Overall, these discoveries will enable further research into the wider biological role of USP7.
Organizational Affiliation:
Almac Discovery Ltd., Centre for Precision Therapeutics, 97 Lisburn Road, Belfast, Northern Ireland BT9 7AE, United Kingdom.