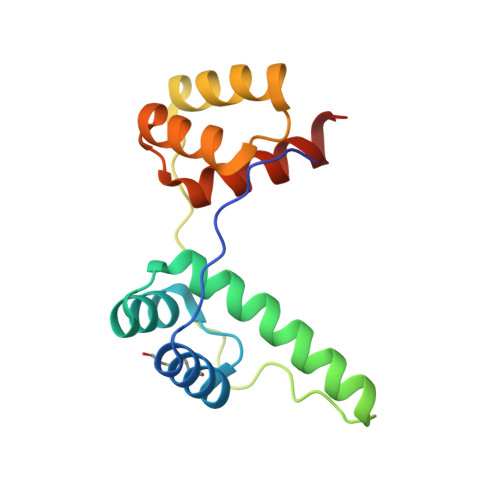
Crystal Structure of the Double Homeodomain of DUX4 in Complex with DNA.
Lee, J.K., Bosnakovski, D., Toso, E.A., Dinh, T., Banerjee, S., Bohl, T.E., Shi, K., Orellana, K., Kyba, M., Aihara, H.(2018) Cell Rep 25: 2955-2962.e3
- PubMed: 30540931
- DOI: https://doi.org/10.1016/j.celrep.2018.11.060
- Primary Citation of Related Structures:
6E8C - PubMed Abstract:
Double homeobox (DUX) transcription factors are unique to eutherian mammals. DUX4 regulates expression of repetitive elements during early embryogenesis, but misexpression of DUX4 causes facioscapulohumeral muscular dystrophy (FSHD) and translocations overexpressing the DUX4 double homeodomain cause B cell leukemia. Here, we report the crystal structure of the tandem homeodomains of DUX4 bound to DNA. The homeodomains bind DNA in a head-to-head fashion, with the linker making anchoring DNA minor-groove interactions and unique protein contacts. Remarkably, despite being tandem duplicates, the DUX4 homeodomains recognize different core sequences. This results from an arginine-to-glutamate mutation, unique to primates, causing alternative positioning of a key arginine side chain in the recognition helix. Mutational studies demonstrate that this primate-specific change is responsible for the divergence in sequence recognition that likely drove coevolution of embryonically regulated repeats in primates. Our work provides a framework for understanding the endogenous function of DUX4 and its role in FSHD and cancer.
Organizational Affiliation:
Department of Biochemistry, Molecular Biology, and Biophysics, University of Minnesota, Minneapolis, MN 55455, USA.