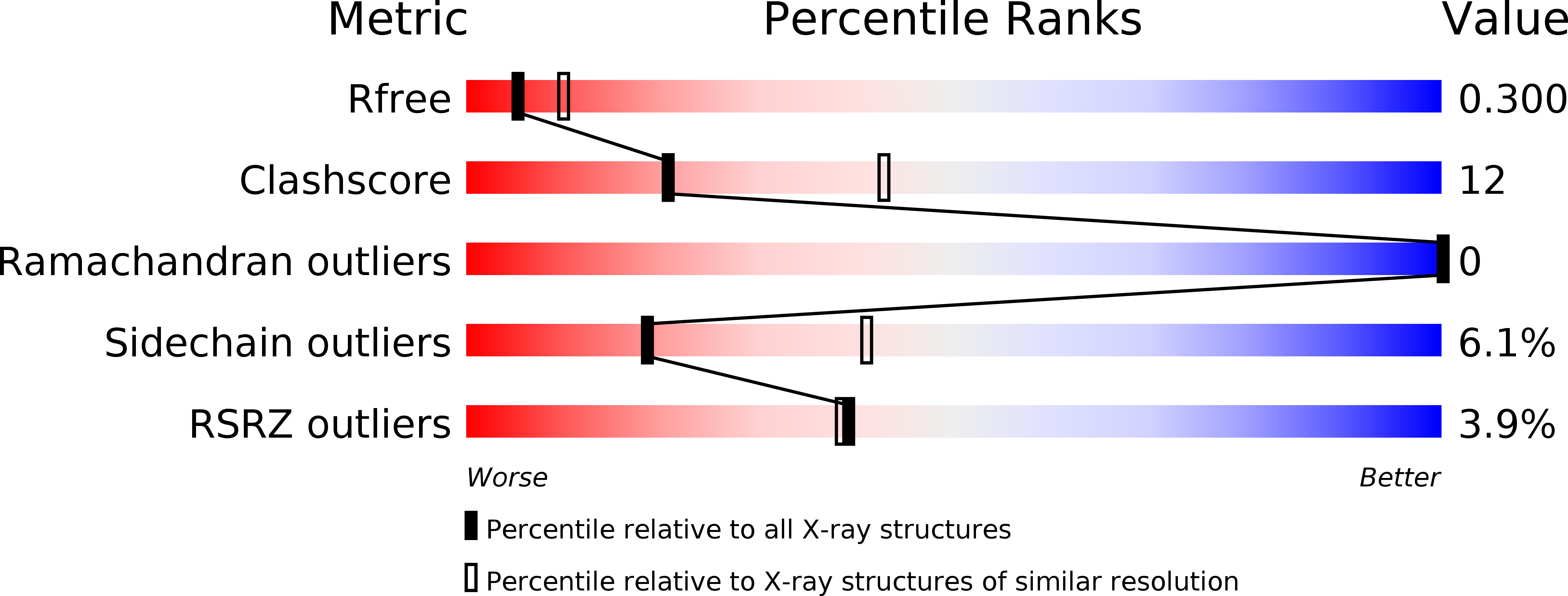
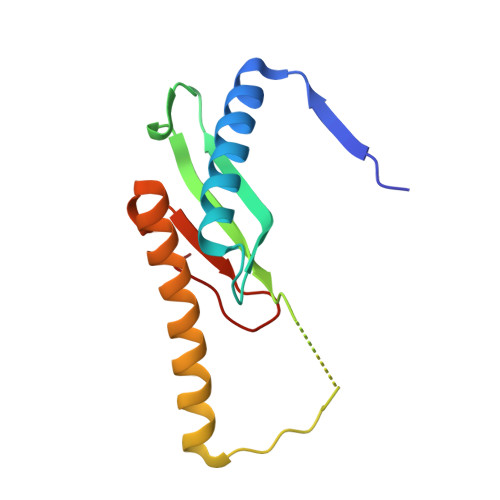
Structural and biochemical characterization of SpoIIIAF, a component of a sporulation-essential channel in Bacillus subtilis.
Zeytuni, N., Flanagan, K.A., Worrall, L.J., Massoni, S.C., Camp, A.H., Strynadka, N.C.J.(2018) J Struct Biol 204: 1-8
- PubMed: 29886194
- DOI: https://doi.org/10.1016/j.jsb.2018.06.002
- Primary Citation of Related Structures:
6DCS - PubMed Abstract:
Environmental stress factors initiate the developmental process of sporulation in some Gram-positive bacteria including Bacillus subtilis. Upon sporulation initiation the bacterial cell undergoes a series of morphological transformations that result in the creation of a single dormant spore. Early in sporulation, an asymmetric cell division produces a larger mother cell and smaller forespore. Next, the mother cell septal membrane engulfs the forespore, and an essential channel, the so-called feeding-tube apparatus, is formed. This assembled channel is thought to form a transenvelope secretion complex that crosses both mother cell and forespore membranes. At least nine proteins are essential for channel formation including SpoIIQ under forespore control and the eight SpoIIIA proteins (SpoIIIAA-AH) under mother cell control. Several of these proteins share similarity with components of Gram-negative bacterial secretion systems and the flagellum. Here we report the X-ray crystallographic structure of the soluble domain of SpoIIIAF to 2.7 Å resolution. Like the channel components SpoIIIAG and SpoIIIAH, SpoIIIAF adopts a conserved ring-building motif (RBM) fold found in proteins from numerous dual membrane secretion systems of distinct function. The SpoIIIAF RBM fold contains two unique features: an extended N-terminal helix, associated with multimerization, and an insertion at a loop region that can adopt two distinct conformations. The ability of the same primary sequence to adopt different secondary structure conformations is associated with protein regulation, suggesting a dual structural and regulatory role for the SpoIIIAF RBM. We further analyzed potential interaction interfaces by structure-guided mutagenesis in vivo. Collectively, our data provide new insight into the possible roles of SpoIIIAF within the secretion-like apparatus during sporulation.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology and the Center for Blood Research, University of British Columbia, Vancouver, British Columbia, Canada.