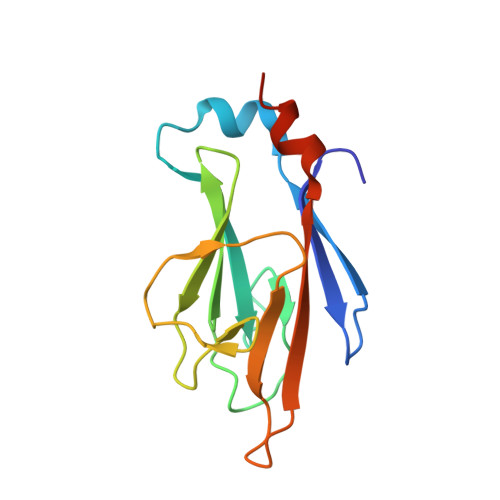
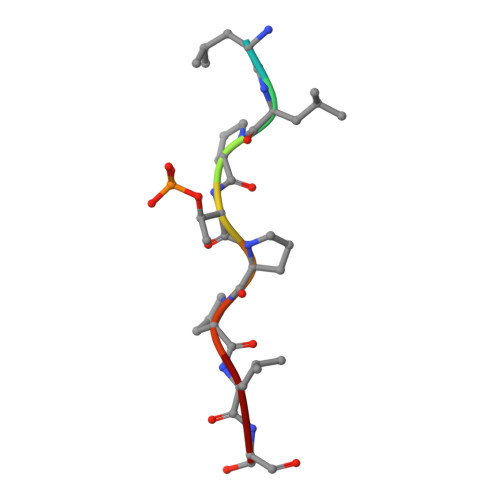
Generating a recombinant phosphothreonine-binding domain for a phosphopeptide of the human transcription factor, c-Myc.
Venegas, L.A., Kall, S.L., Bankole, O., Lavie, A., Kay, B.K.(2018) N Biotechnol 45: 36-44
- PubMed: 29763736
- DOI: https://doi.org/10.1016/j.nbt.2018.05.001
- Primary Citation of Related Structures:
6C4U - PubMed Abstract:
Transcription factor c-Myc is an oncoprotein that is regulated at the post-translational level through phosphorylation of two conserved residues, Serine 62 (Ser62) and Threonine 58 (Thr58). A highly specific tool capable of recognizing Myc via pThr58 is needed to monitor activation and localization. Through phage display, we have isolated 10 engineered Forkhead-associated (FHA) domains that selectively bind to a phosphothreonine (pThr)-containing peptide (53-FELLPpTPPLSPS-64) segment of human c-Myc. One domain variant was observed to bind to the Myc-pThr58 peptide with a K D value of 800 nM and had >1000-fold discrimination between the phosphorylated and non-phosphorylated peptide. The crystal structure of the engineered FHA Myc-pThr-binding domain (Myc-pTBD) was solved in complex with its cognate ligand. The Myc-pTBD was observed to be structurally similar to the yeast Rad9 FHA1 domain, except that its β4-β5 and β10-β11 loops form a hydrophobic pocket to facilitate the interaction between the domain and the peptide ligand. The Myc-pTBD's specificity for its cognate ligand was demonstrated to be on a par with 3 commercial polyclonal antibodies, suggesting that this recombinant reagent is a viable alternative to antibodies for monitoring Myc regulation.
Organizational Affiliation:
Department of Biological Sciences, University of Illinois at Chicago, Chicago, IL 60607, USA.