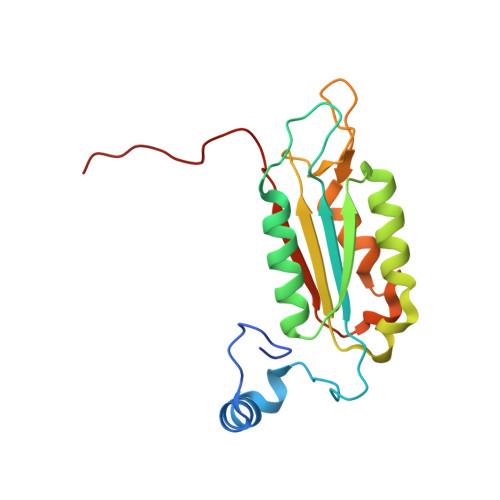
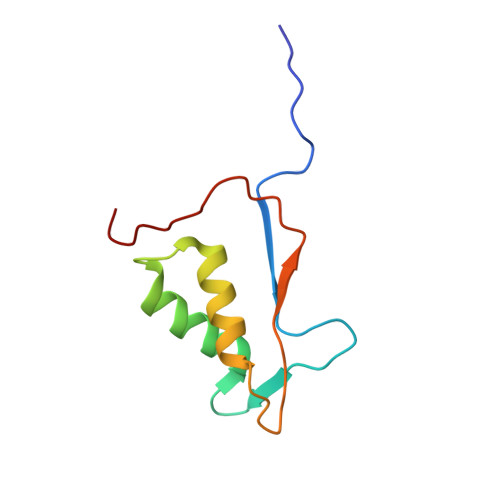
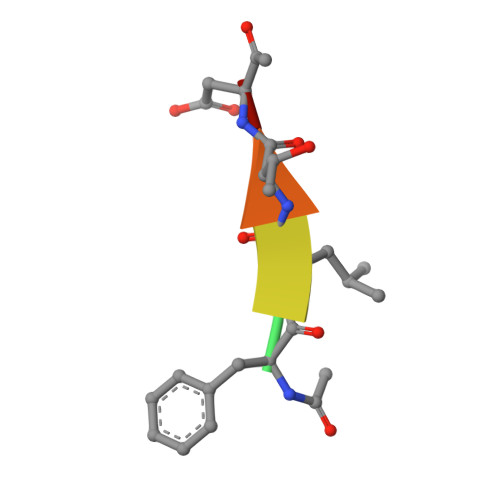
Mechanism of gasdermin D recognition by inflammatory caspases and their inhibition by a gasdermin D-derived peptide inhibitor.
Yang, J., Liu, Z., Wang, C., Yang, R., Rathkey, J.K., Pinkard, O.W., Shi, W., Chen, Y., Dubyak, G.R., Abbott, D.W., Xiao, T.S.(2018) Proc Natl Acad Sci U S A 115: 6792-6797
- PubMed: 29891674
- DOI: https://doi.org/10.1073/pnas.1800562115
- Primary Citation of Related Structures:
6BZ9 - PubMed Abstract:
The inflammasomes are signaling platforms that promote the activation of inflammatory caspases such as caspases-1, -4, -5, and -11. Recent studies identified gasdermin D (GSDMD) as an effector for pyroptosis downstream of the inflammasome signaling pathways. Cleavage of GSDMD by inflammatory caspases allows its N-terminal domain to associate with membrane lipids and form pores that induce pyroptotic cell death. Despite the important role of GSDMD in pyroptosis, the molecular mechanisms of GSDMD recognition and cleavage by inflammatory caspases that trigger pyroptosis are poorly understood. Here, we demonstrate that the catalytic domains of inflammatory caspases can directly bind to both the full-length GSDMD and its cleavage site peptide, FLTD. A GSDMD-derived inhibitor, N -acetyl-Phe-Leu-Thr-Asp-chloromethylketone (Ac-FLTD-CMK), inhibits GSDMD cleavage by caspases-1, -4, -5, and -11 in vitro, suppresses pyroptosis downstream of both canonical and noncanonical inflammasomes, as well as reduces IL-1β release following activation of the NLRP3 inflammasome in macrophages. By contrast, the inhibitor does not target caspase-3 or apoptotic cell death, suggesting that Ac-FLTD-CMK is a specific inhibitor for inflammatory caspases. Crystal structure of caspase-1 in complex with Ac-FLTD-CMK reveals extensive enzyme-inhibitor interactions involving both hydrogen bonds and hydrophobic contacts. Comparison with other caspase-1 structures demonstrates drastic conformational changes at the four active-site loops that assemble the catalytic groove. The present study not only contributes to our understanding of GSDMD recognition by inflammatory caspases but also reports a specific inhibitor for these caspases that can serve as a tool for investigating inflammasome signaling.
Organizational Affiliation:
Department of Pathology, Case Western Reserve University School of Medicine, Cleveland, OH 44106.