Structural insights into IFIT1 dimerization and conformational changes associated with mRNA binding
Abbas, Y.M., Martinez-Montero, S., Cencic, R., Pelletier, J., Damha, M.J., Pawelek, P.D., Nagar, B.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Interferon-induced protein with tetratricopeptide repeats 1 | 479 | Homo sapiens | Mutation(s): 0 Gene Names: IFIT1, G10P1, IFI56, IFNAI1, ISG56 | 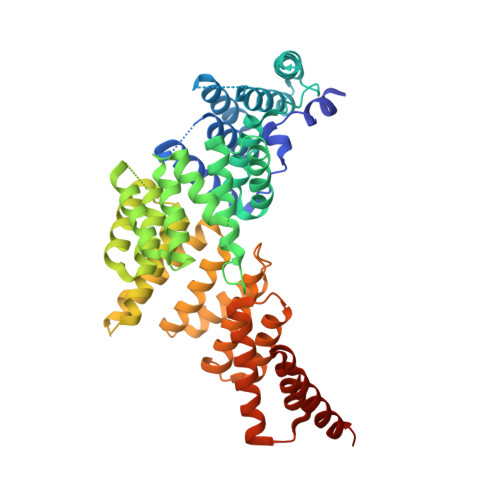 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P09914 (Homo sapiens) Explore P09914 Go to UniProtKB: P09914 | |||||
PHAROS: P09914 GTEx: ENSG00000185745 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P09914 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| RNA (5'-D(*(ATP))-R(P*AP*AP*A)-3') | 4 | unidentified | 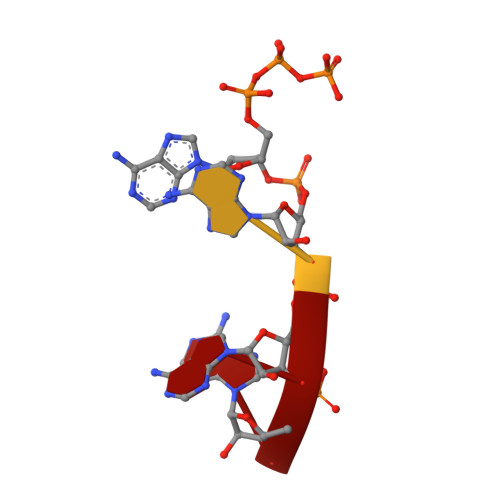 | ||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 184.789 | α = 90 |
| b = 184.789 | β = 90 |
| c = 88.864 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data scaling |
| PHASER | phasing |
| Coot | model building |