Targeting lysine specific demethylase 4A (KDM4A) tandem TUDOR domain - A fragment based approach.
Upadhyay, A.K., Judge, R.A., Li, L., Pithawalla, R., Simanis, J., Bodelle, P.M., Marin, V.L., Henry, R.F., Petros, A.M., Sun, C.(2018) Bioorg Med Chem Lett 28: 1708-1713
- PubMed: 29691138
- DOI: https://doi.org/10.1016/j.bmcl.2018.04.050
- Primary Citation of Related Structures:
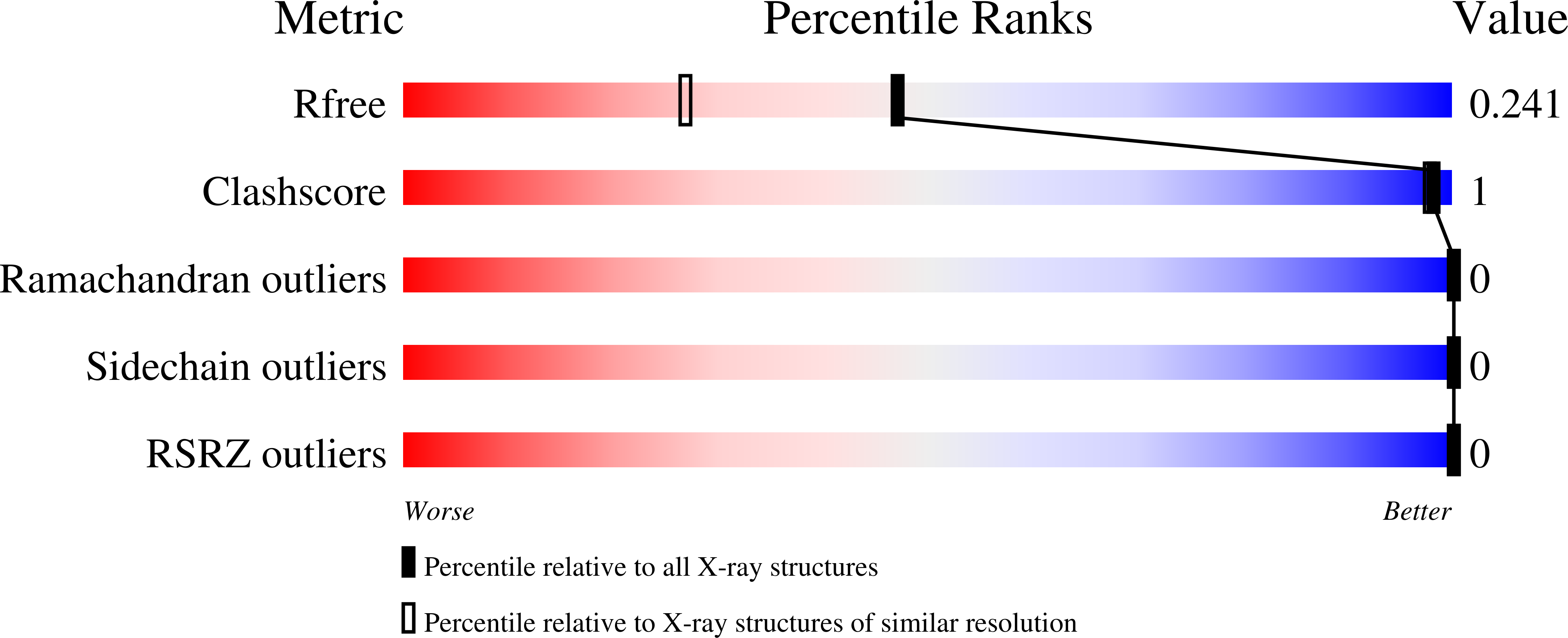
5VAR - PubMed Abstract:
The tandem TUDOR domains present in the non-catalytic C-terminal half of the KDM4A, 4B and 4C enzymes play important roles in regulating their chromatin localizations and substrate specificities. They achieve this regulatory role by binding to different tri-methylated lysine residues on histone H3 (H3-K4me3, H3-K23me3) and histone H4 (H4-K20me3) depending upon the specific chromatin environment. In this work, we have used a 2D-NMR based fragment screening approach to identify a novel fragment (1a), which binds to the KDM4A-TUDOR domain and shows modest competition with H3-K4me3 binding in biochemical as well as in vitro cell based assays. A co-crystal structure of KDM4A TUDOR domain in complex with 1a shows that the fragment binds stereo-specifically to the methyl lysine binding pocket forming a network of strong hydrogen bonds and hydrophobic interactions. We anticipate that the fragment 1a can be further developed into a novel allosteric inhibitor of the KDM4 family of enzymes through targeting their C-terminal tandem TUDOR domain.
Organizational Affiliation:
AbbVie Inc., 1 North Waukegan Road, North Chicago, IL 60064, USA. Electronic address: anup.upadhyay@abbvie.com.