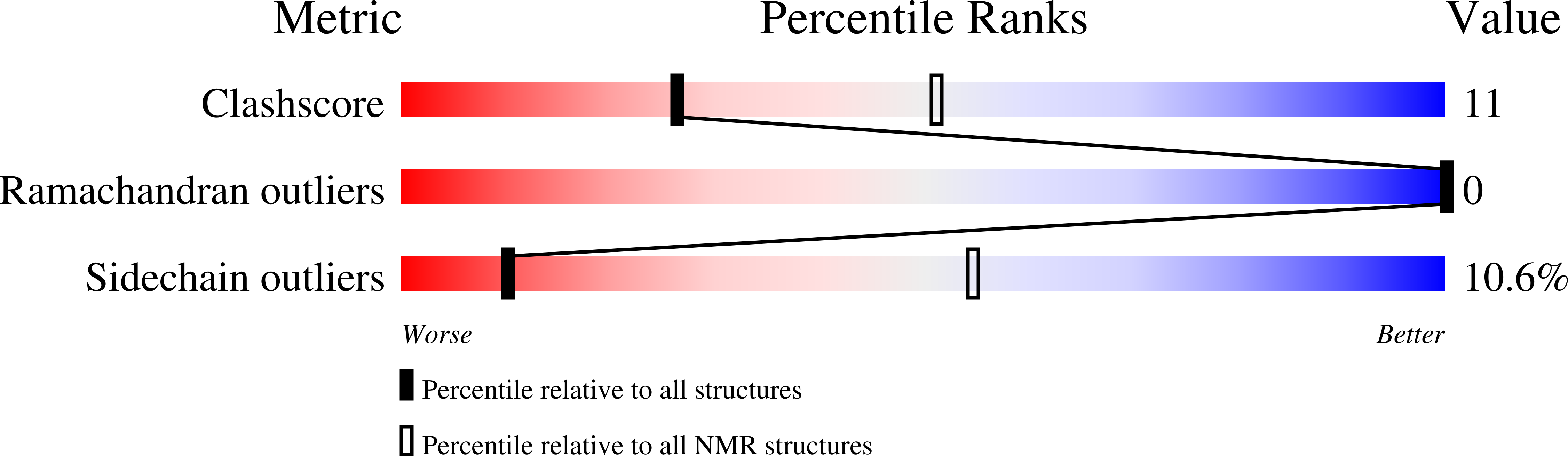The Conformation of the Epidermal Growth Factor Receptor Transmembrane Domain Dimer Dynamically Adapts to the Local Membrane Environment.
Bocharov, E.V., Bragin, P.E., Pavlov, K.V., Bocharova, O.V., Mineev, K.S., Polyansky, A.A., Volynsky, P.E., Efremov, R.G., Arseniev, A.S.(2017) Biochemistry 56: 1697-1705
- PubMed: 28291355
- DOI: https://doi.org/10.1021/acs.biochem.6b01085
- Primary Citation of Related Structures:
5LV6 - PubMed Abstract:
The epidermal growth factor receptor (EGFR) family is an important class of receptor tyrosine kinases, mediating a variety of cellular responses in normal biological processes and in pathological states of multicellular organisms. Different modes of dimerization of the human EGFR transmembrane domain (TMD) in different membrane mimetics recently prompted us to propose a novel signal transduction mechanism based on protein-lipid interaction. However, the experimental evidence for it was originally obtained with slightly different TMD fragments used in the two different mimetics, compromising the validity of the comparison. To eliminate ambiguity, we determined the nuclear magnetic resonance (NMR) structure of the bicelle-incorporated dimer of the EGFR TMD fragment identical to the one previously used in micelles. The NMR results augmented by molecular dynamics simulations confirm the mutual influence of the TMD and lipid environment, as is required for the proposed lipid-mediated activation mechanism. They also reveal the possible functional relevance of a subtle interplay between the concurrent processes in the lipid and protein during signal transduction.
Organizational Affiliation:
Department of Structural Biology, Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry RAS , str. Miklukho-Maklaya 16/10, Moscow 117997, Russian Federation.