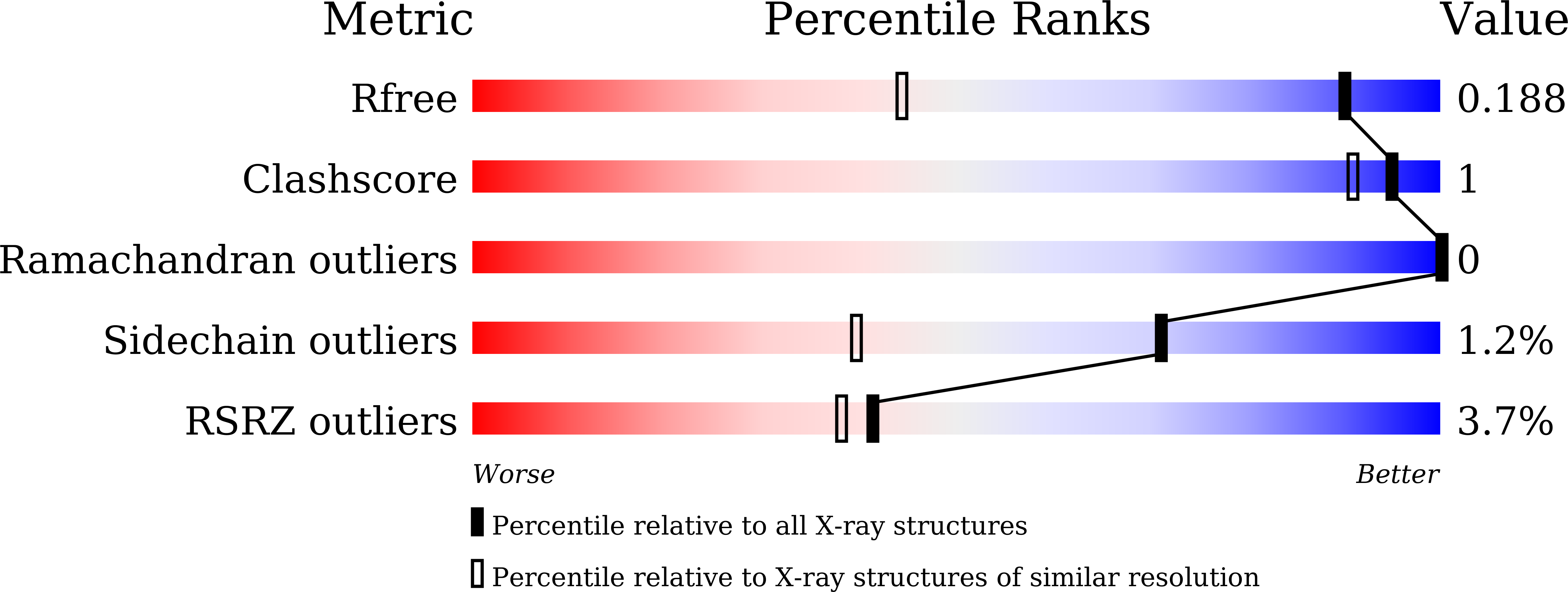
RCC2 is a novel p53 target in suppressing metastasis.
Song, C., Liang, L., Jin, Y., Li, Y., Liu, Y., Guo, L., Wu, C., Yun, C.H., Yin, Y.(2018) Oncogene 37: 8-17
- PubMed: 28869598
- DOI: https://doi.org/10.1038/onc.2017.306
- Primary Citation of Related Structures:
5GWN - PubMed Abstract:
RCC2 (also known as TD60) is a highly conserved protein involved in prognosis in colorectal cancer. However, its relationship with tumor development is less understood. Here we demonstrate a signaling pathway defining regulation of RCC2 and its functions in tumor progression. We report that p53 is a transcriptional regulator of RCC2 that acts through its binding to a palindromic motif in the RCC2 promoter. RCC2 physically interacts and deactivates a small GTPase Rac1 that is known to be involved in metastasis. We solved a high-resolution crystal structure of RCC2 and revealed one RCC1-like domain with a unique β-hairpin that is requisite for RCC2 interaction with Rac1. p53 or RCC2 deficiency leads to activation of Rac1 and deterioration of extracellular matrix sensing (haptotaxis) of surface-bound gradients. Ectopic expression of RCC2 restores directional migration in p53-null cells. Our results demonstrate that p53 and RCC2 signaling is important for regulation of cell migration and suppression of metastasis. We propose that the p53/RCC2/Rac1 axis is a potential target for cancer therapy.
Organizational Affiliation:
Institute of Systems Biomedicine, Departments of Pathology and Biophysics, School of Basic Medical Sciences, Beijing Key Laboratory of Tumor Systems Biology, Peking-Tsinghua Center for Life Sciences, Peking University Health Science Center, Beijing, China.