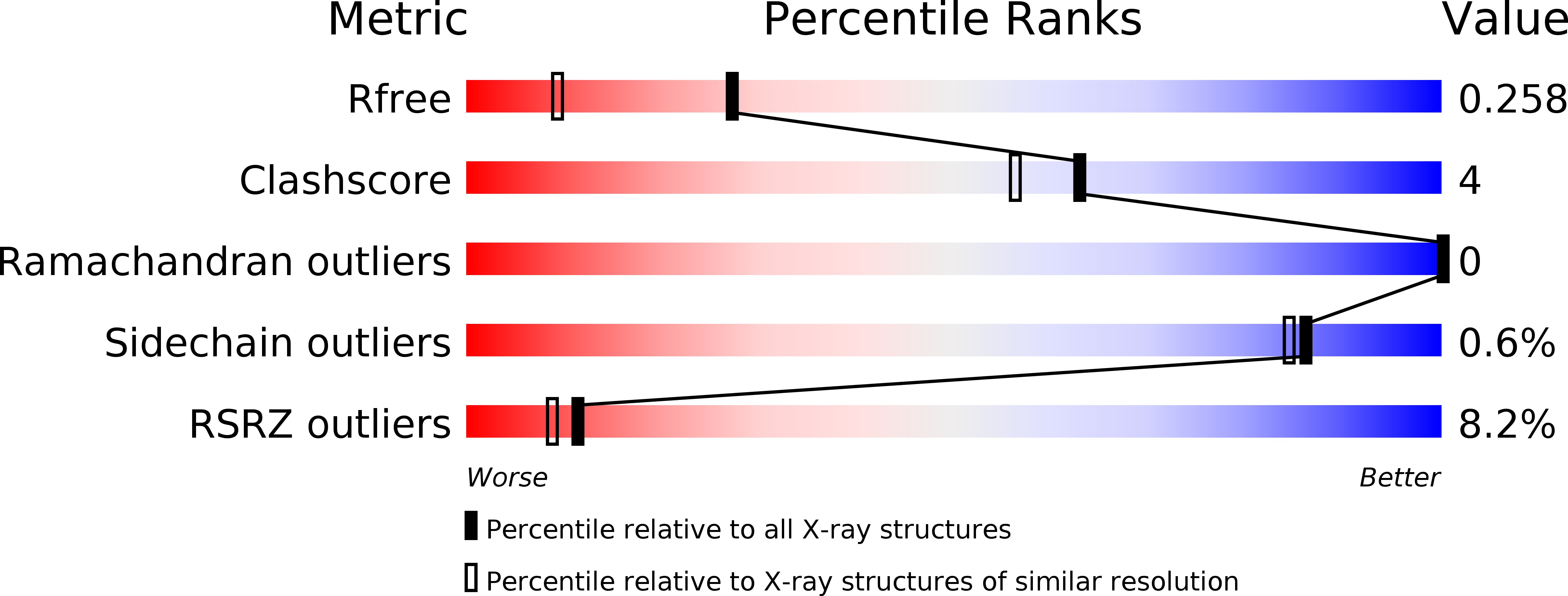
An enantioselective artificial Suzukiase based on the biotin-streptavidin technology.
Chatterjee, A., Mallin, H., Klehr, J., Vallapurackal, J., Finke, A.D., Vera, L., Marsh, M., Ward, T.R.(2016) Chem Sci 7: 673-677
- PubMed: 29896353
- DOI: https://doi.org/10.1039/c5sc03116h
- Primary Citation of Related Structures:
5CSE - PubMed Abstract:
Introduction of a biotinylated monophosphine palladium complex within streptavidin affords an enantioselective artificial Suzukiase. Site-directed mutagenesis allowed the optimization of the activity and the enantioselectivity of this artificial metalloenzyme. A variety of atropisomeric biaryls were produced in good yields and up to 90% ee. The hybrid catalyst described herein shows comparable TOF to the previous aqueous-asymmetric Suzuki catalysts, and excellent stability under the reaction conditions to realize higher TON through longer reaction time.
Organizational Affiliation:
Department of Chemistry , University of Basel , Spitalstrasse 51 , 4056 Basel , Switzerland . Email: thomas.ward@unibas.ch.