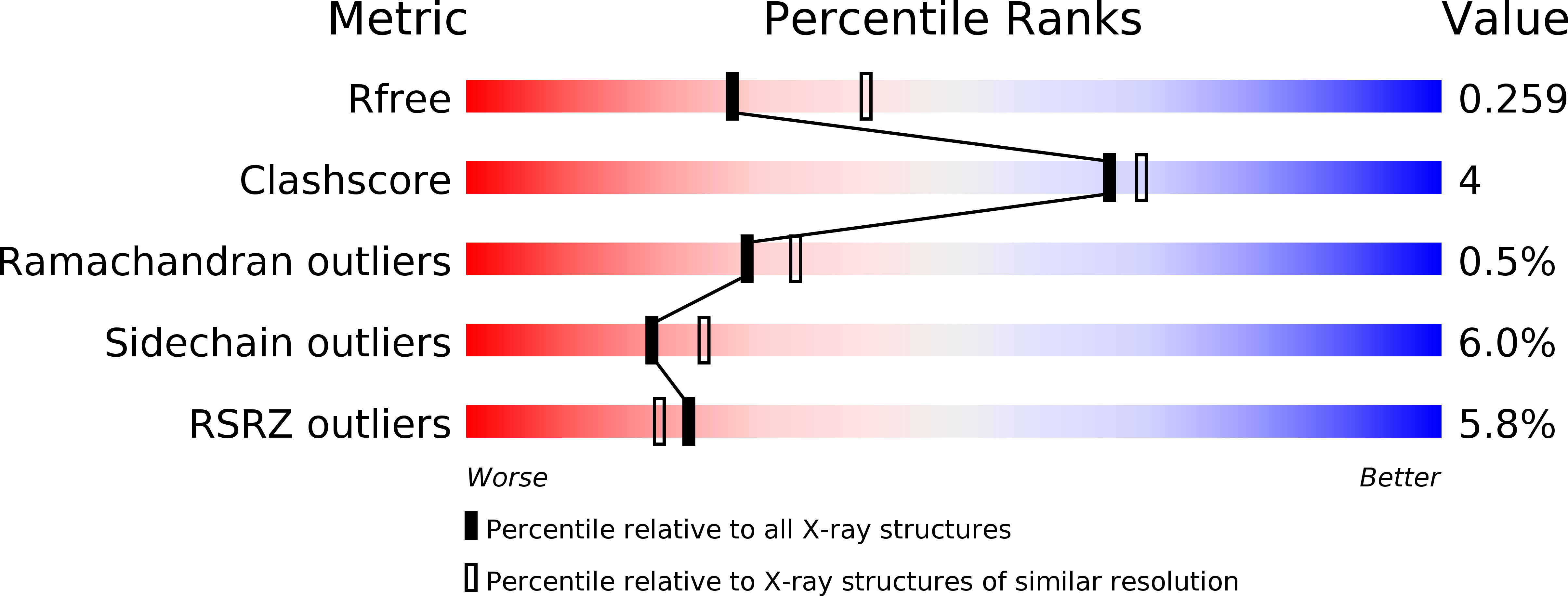
Fluorescein Derivatives as Bifunctional Molecules for the Simultaneous Inhibiting and Labeling of FTO Protein
Wang, T., Hong, T., Huang, Y., Su, H., Wu, F., Chen, Y., Wei, L., Huang, W., Hua, X., Xia, Y., Xu, J., Gan, J., Yuan, B., Feng, Y., Zhang, X., Yang, C.G., Zhou, X.(2015) J Am Chem Soc 137: 13736-13739
- PubMed: 26457839
- DOI: https://doi.org/10.1021/jacs.5b06690
- Primary Citation of Related Structures:
4ZS2, 4ZS3 - PubMed Abstract:
The FTO protein is unequivocally reported to play a critical role in human obesity and in the regulation of cellular levels of m(6)A modification, which makes FTO a significant and worthy subject of study. Here, we identified that fluorescein derivatives can selectively inhibit FTO demethylation, and the mechanisms behind these activities were elucidated after we determined the X-ray crystal structures of FTO/fluorescein and FTO/5-aminofluorescein. Furthermore, these inhibitors can also be applied to the direct labeling and enrichment of FTO protein combined with photoaffinity labeling assay.
Organizational Affiliation:
State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences , Shanghai 201203, China.