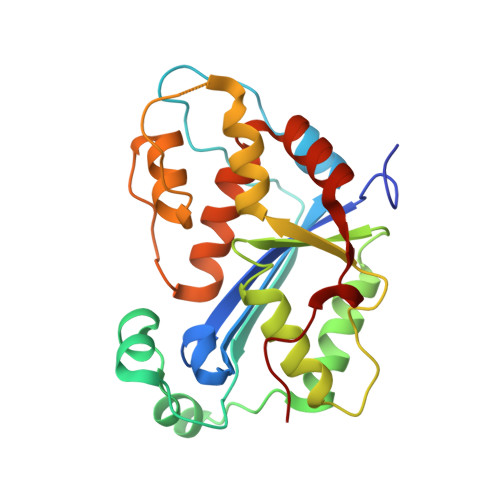
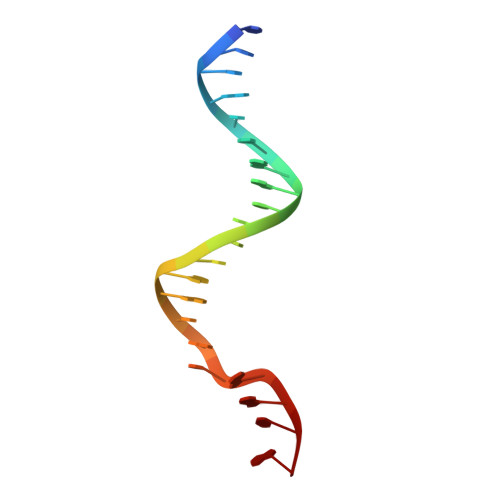
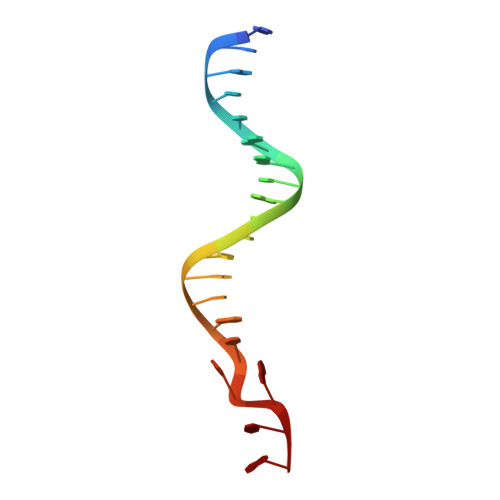
Exonuclease TREX1 degrades double-stranded DNA to prevent spontaneous lupus-like inflammatory disease.
Grieves, J.L., Fye, J.M., Harvey, S., Grayson, J.M., Hollis, T., Perrino, F.W.(2015) Proc Natl Acad Sci U S A 112: 5117-5122
- PubMed: 25848017
- DOI: https://doi.org/10.1073/pnas.1423804112
- Primary Citation of Related Structures:
4YNQ - PubMed Abstract:
The TREX1 gene encodes a potent DNA exonuclease, and mutations in TREX1 cause a spectrum of lupus-like autoimmune diseases. Most lupus patients develop autoantibodies to double-stranded DNA (dsDNA), but the source of DNA antigen is unknown. The TREX1 D18N mutation causes a monogenic, cutaneous form of lupus called familial chilblain lupus, and the TREX1 D18N enzyme exhibits dysfunctional dsDNA-degrading activity, providing a link between dsDNA degradation and nucleic acid-mediated autoimmune disease. We determined the structure of the TREX1 D18N protein in complex with dsDNA, revealing how this exonuclease uses a novel DNA-unwinding mechanism to separate the polynucleotide strands for single-stranded DNA (ssDNA) loading into the active site. The TREX1 D18N dsDNA interactions coupled with catalytic deficiency explain how this mutant nuclease prevents dsDNA degradation. We tested the effects of TREX1 D18N in vivo by replacing the TREX1 WT gene in mice with the TREX1 D18N allele. The TREX1 D18N mice exhibit systemic inflammation, lymphoid hyperplasia, vasculitis, and kidney disease. The observed lupus-like inflammatory disease is associated with immune activation, production of autoantibodies to dsDNA, and deposition of immune complexes in the kidney. Thus, dysfunctional dsDNA degradation by TREX1 D18N induces disease in mice that recapitulates many characteristics of human lupus. Failure to clear DNA has long been linked to lupus in humans, and these data point to dsDNA as a key substrate for TREX1 and a major antigen source in mice with dysfunctional TREX1 enzyme.
Organizational Affiliation:
Department of Biochemistry.