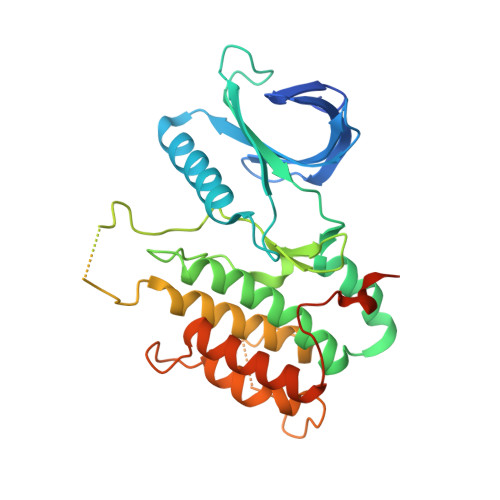
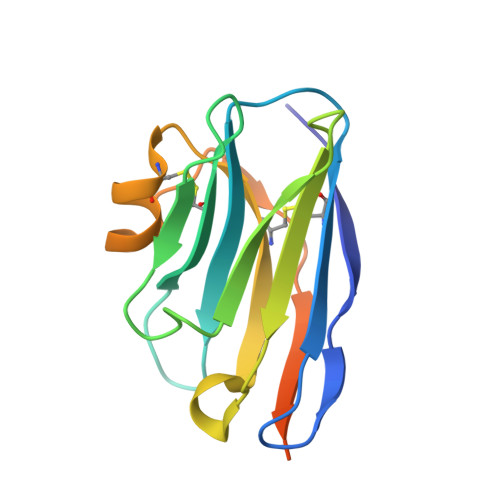
Selective Inhibitors of Cyclin G Associated Kinase (GAK) as Anti-Hepatitis C Agents.
Kovackova, S., Chang, L., Bekerman, E., Neveu, G., Barouch-Bentov, R., Chaikuad, A., Heroven, C., Sala, M., De Jonghe, S., Knapp, S., Einav, S., Herdewijn, P.(2015) J Med Chem 58: 3393-3410
- PubMed: 25822739
- DOI: https://doi.org/10.1021/jm501759m
- Primary Citation of Related Structures:
4Y8D - PubMed Abstract:
Cyclin G associated kinase (GAK) emerged as a promising drug target for the treatment of viral infections. However, no potent and selective GAK inhibitors have been reported in the literature to date. This paper describes the discovery of isothiazolo[5,4-b]pyridines as selective GAK inhibitors, with the most potent congeners displaying low nanomolar binding affinity for GAK. Cocrystallization experiments revealed that these compounds behaved as classic type I ATP-competitive kinase inhibitors. In addition, we have demonstrated that these compounds exhibit a potent activity against hepatitis C virus (HCV) by inhibiting two temporally distinct steps in the HCV life cycle (i.e., viral entry and assembly). Hence, these GAK inhibitors represent chemical probes to study GAK function in different disease areas where GAK has been implicated (including viral infection, cancer, and Parkinson's disease).
Organizational Affiliation:
†Laboratory of Medicinal Chemistry, Rega Institute for Medical Research, KU Leuven, Minderbroedersstraat 10, 3000 Leuven, Belgium.