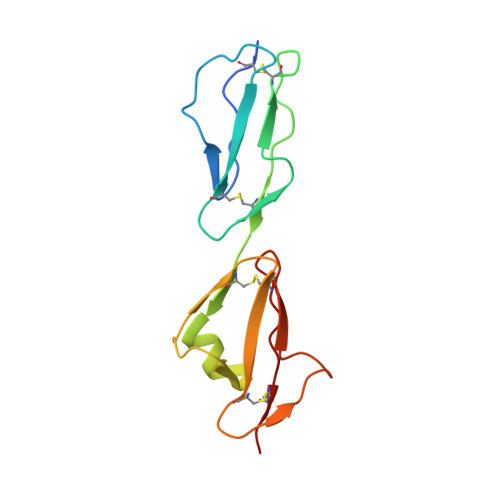
Structural basis for sialic acid-mediated self-recognition by complement factor H.
Blaum, B.S., Hannan, J.P., Herbert, A.P., Kavanagh, D., Uhrin, D., Stehle, T.(2015) Nat Chem Biol 11: 77-82
- PubMed: 25402769
- DOI: https://doi.org/10.1038/nchembio.1696
- Primary Citation of Related Structures:
4ONT - PubMed Abstract:
The serum protein complement factor H (FH) ensures downregulation of the complement alternative pathway, a branch of innate immunity, upon interaction with specific glycans on host cell surfaces. Using ligand-based NMR, we screened a comprehensive set of sialylated glycans for binding to FH and solved the crystal structure of a ternary complex formed by the two C-terminal domains of FH, a sialylated trisaccharide and the complement C3b thioester-containing domain. Key residues in the sialic acid binding site are conserved from mice to men, and residues linked to atypical hemolytic uremic syndrome cluster within this binding site, suggesting a possible role for sialic acid as a host marker also in other mammals and a critical role in human renal complement homeostasis. Unexpectedly, the FH sialic acid binding site is structurally homologous to the binding sites of two evolutionarily unrelated proteins. The crystal structure also advances our understanding of bacterial immune evasion strategies.
Organizational Affiliation:
Interfaculty Institute of Biochemistry, University of Tübingen, Tübingen, Germany.