Structural, Kinetic, and Docking Studies of Artificial Imine Reductases Based on Biotin-Streptavidin Technology: An Induced Lock-and-Key Hypothesis
Robles, V.M., Durrenberger, M., Heinisch, T., Lledos, A., Schirmer, T., Ward, T.R., Marechal, J.D.(2014) J Am Chem Soc 136: 15676-15683
- PubMed: 25317660
- DOI: https://doi.org/10.1021/ja508258t
- Primary Citation of Related Structures:
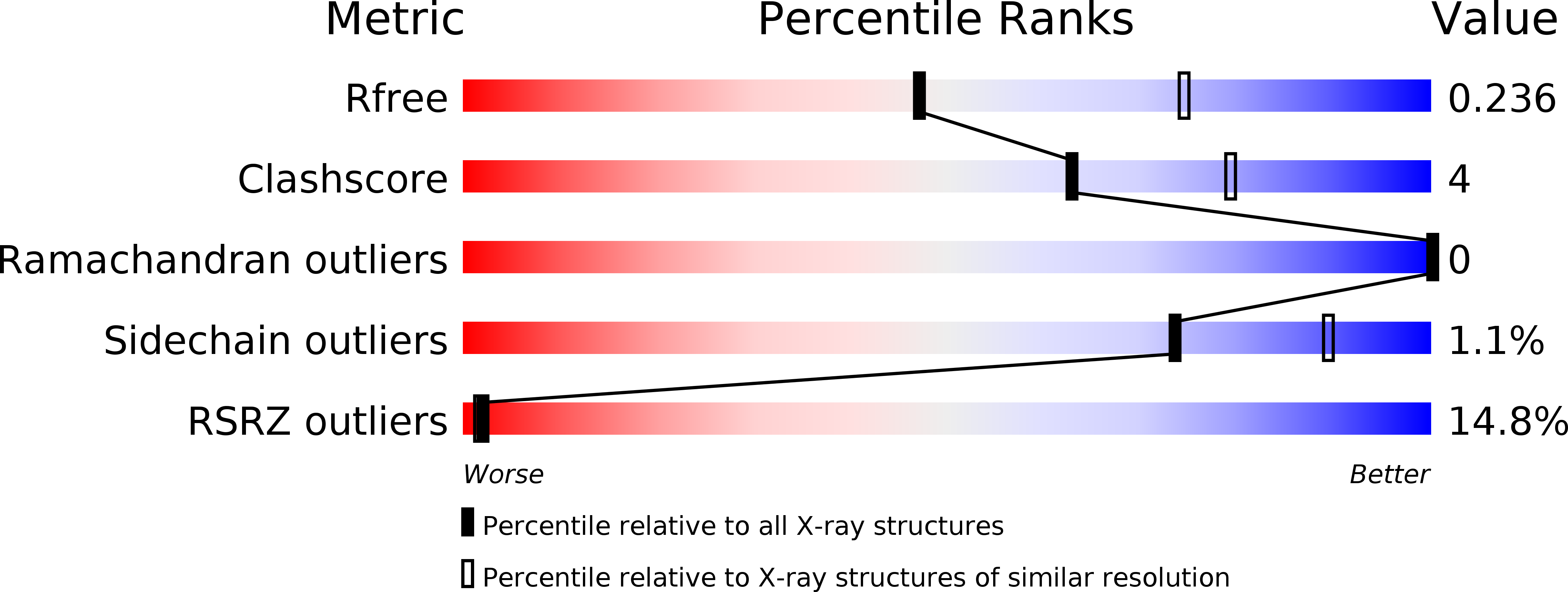
4OKA - PubMed Abstract:
An artificial imine reductase results upon incorporation of a biotinylated Cp*Ir moiety (Cp* = C5Me5(-)) within homotetrameric streptavidin (Sav) (referred to as Cp*Ir(Biot-p-L)Cl] ⊂ Sav). Mutation of S112 reveals a marked effect of the Ir/streptavidin ratio on both the saturation kinetics as well as the enantioselectivity for the production of salsolidine. For [Cp*Ir(Biot-p-L)Cl] ⊂ S112A Sav, both the reaction rate and the selectivity (up to 96% ee (R)-salsolidine, kcat 14-4 min(-1) vs [Ir], KM 65-370 mM) decrease upon fully saturating all biotin binding sites (the ee varying between 96% ee and 45% ee R). In contrast, for [Cp*Ir(Biot-p-L)Cl] ⊂ S112K Sav, both the rate and the selectivity remain nearly constant upon varying the Ir/streptavidin ratio [up to 78% ee (S)-salsolidine, kcat 2.6 min(-1), KM 95 mM]. X-ray analysis complemented with docking studies highlight a marked preference of the S112A and S112K Sav mutants for the SIr and RIr enantiomeric forms of the cofactor, respectively. Combining both docking and saturation kinetic studies led to the formulation of an enantioselection mechanism relying on an "induced lock-and-key" hypothesis: the host protein dictates the configuration of the biotinylated Ir-cofactor which, in turn, by and large determines the enantioselectivity of the imine reductase.
Organizational Affiliation:
Departament de Química, Universitat Autònoma de Barcelona , Edifici C.n., 08193 Cerdanyola del Vallés, Barcelona, Spain.