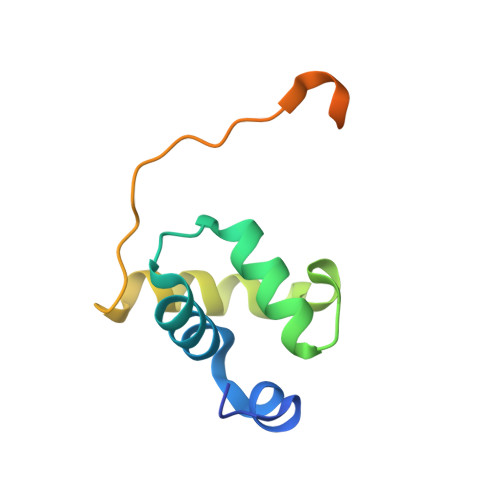
X-ray vs. NMR structure of N-terminal domain of delta-subunit of RNA polymerase.
Demo, G., Papouskova, V., Komarek, J., Kaderavek, P., Otrusinova, O., Srb, P., Rabatinova, A., Krasny, L., Zidek, L., Sklenar, V., Wimmerova, M.(2014) J Struct Biol 187: 174-186
- PubMed: 24937760
- DOI: https://doi.org/10.1016/j.jsb.2014.06.001
- Primary Citation of Related Structures:
4NC7, 4NC8 - PubMed Abstract:
The crystal structure of the N-terminal domain of the RNA polymerase δ subunit (Nδ) from Bacillus subtilis solved at a resolution of 2.0Å is compared with the NMR structure determined previously. The molecule crystallizes in the space group C222(1) with a dimer in the asymmetric unit. Importantly, the X-ray structure exhibits significant differences from the lowest energy NMR structure. In addition to the overall structure differences, structurally important β sheets found in the NMR structure are not present in the crystal structure. We systematically investigated the cause of the discrepancies between the NMR and X-ray structures of Nδ, addressing the pH dependence, presence of metal ions, and crystal packing forces. We convincingly showed that the crystal packing forces, together with the presence of Ni(2+) ions, are the main reason for such a difference. In summary, the study illustrates that the two structural approaches may give unequal results, which need to be interpreted with care to obtain reliable structural information in terms of biological relevance.
Organizational Affiliation:
National Centre for Biomolecular Research, Faculty of Science, Masaryk University, Kamenice 5/A4, 62500 Brno, Czech Republic; Central European Institute of Technology-CEITEC, Masaryk University, Kamenice 5/A4, 62500 Brno, Czech Republic.