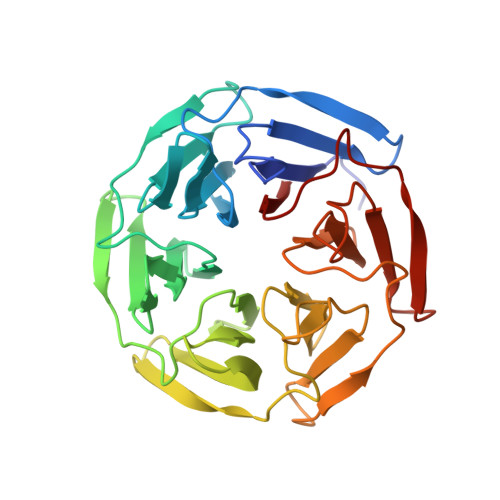
Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes
Pan, H., Lin, M., Yang, Y., Callaway, K., Baker, J., Diep, L., Yan, J., Tanaka, K., Zhu, Y.L., Konradi, A.W., Jobling, M., Tam, D., Ren, Z., Cheung, H., Bova, M., Riley, B.E., Yao, N., Artis, D.R.To be published.