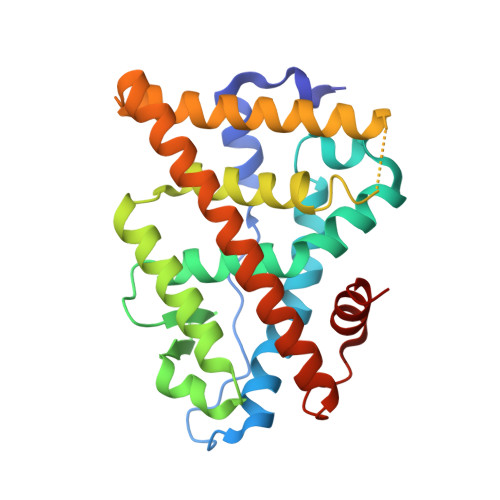
Structural basis for a molecular allosteric control mechanism of cofactor binding to nuclear receptors.
Osz, J., Brelivet, Y., Peluso-Iltis, C., Cura, V., Eiler, S., Ruff, M., Bourguet, W., Rochel, N., Moras, D.(2012) Proc Natl Acad Sci U S A 109: E588-E594
- PubMed: 22355136
- DOI: https://doi.org/10.1073/pnas.1118192109
- Primary Citation of Related Structures:
4DM6, 4DM8, 4DMA - PubMed Abstract:
Transcription regulation by steroid hormones, vitamin derivatives, and metabolites is mediated by nuclear receptors (NRs), which play an important role in ligand-dependent gene expression and human health. NRs function as homodimers or heterodimers and are involved in a combinatorial, coordinated and sequentially orchestrated exchange between coregulators (corepressors, coactivators). The architecture of DNA-bound functional dimers positions the coregulators proteins. We previously demonstrated that retinoic acid (RAR-RXR) and vitamin D3 receptors (VDR-RXR) heterodimers recruit only one coactivator molecule asymmetrically without steric hindrance for the binding of a second cofactor. We now address the problem of homodimers for which the presence of two identical targets enhances the functional importance of the mode of binding. Using structural and biophysical methods and RAR as a model, we could dissect the molecular mechanism of coactivator recruitment to homodimers. Our study reveals an allosteric mechanism whereby binding of a coactivator promotes formation of nonsymmetrical RAR homodimers with a 21 stoichiometry. Ligand conformation and the cofactor binding site of the unbound receptor are affected through the dimer interface. A similar control mechanism is observed with estrogen receptor (ER) thus validating the negative cooperativity model for an established functional homodimer. Correlation with published data on other NRs confirms the general character of this regulatory pathway.
Organizational Affiliation:
Institut National de Santé et de Recherche Médicale U964/Centre National de Recherche Scientifique, Unité Mixte de Recherche 7104/Université de Strasbourg, 67404 Illkirch, France.