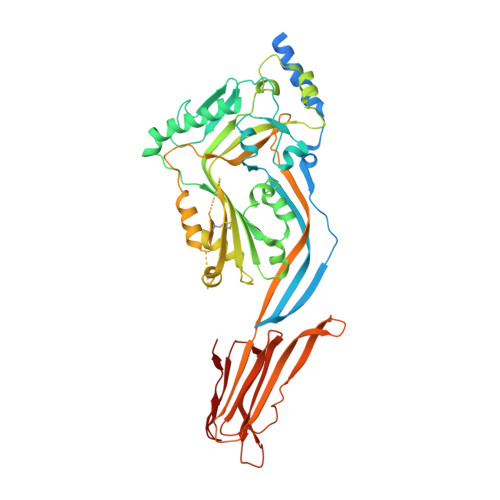
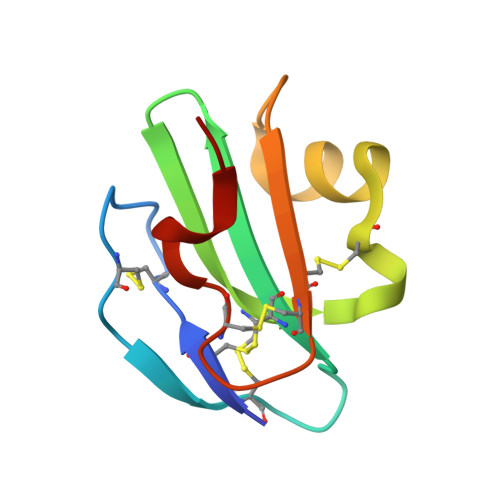
Structural Basis for Recognition of the Pore- Forming Toxin Intermedilysin by Human Complement Receptor Cd59
Johnson, S., Brooks, N.J., Smith, R.A.G., Lea, S.M., Bubeck, D.(2013) Cell Rep 3: 1369
- PubMed: 23665225
- DOI: https://doi.org/10.1016/j.celrep.2013.04.029
- Primary Citation of Related Structures:
4BIK - PubMed Abstract:
Pore-forming proteins containing the structurally conserved membrane attack complex/perforin fold play an important role in immunity and host-pathogen interactions. Intermedilysin (ILY) is an archetypal member of a cholesterol-dependent cytolysin subclass that hijacks the complement receptor CD59 to make cytotoxic pores in human cells. ILY directly competes for the membrane attack complex binding site on CD59, rendering cells susceptible to complement lysis. To understand how these bacterial pores form in lipid bilayers and the role CD59 plays in complement regulation, we determined the crystal structure of human CD59 bound to ILY. Here, we show the ILY-CD59 complex at 3.5 Å resolution and identify two interfaces mediating this host-pathogen interaction. An ILY-derived peptide based on the binding site inhibits pore formation in a CD59-containing liposome model system. These data provide insight into how CD59 coordinates ILY monomers, nucleating an early prepore state, and suggest a potential mechanism of inhibition for the complement terminal pathway.
Organizational Affiliation:
Sir William Dunn School of Pathology, University of Oxford, South Parks Road, Oxford OX1 3RE, UK.