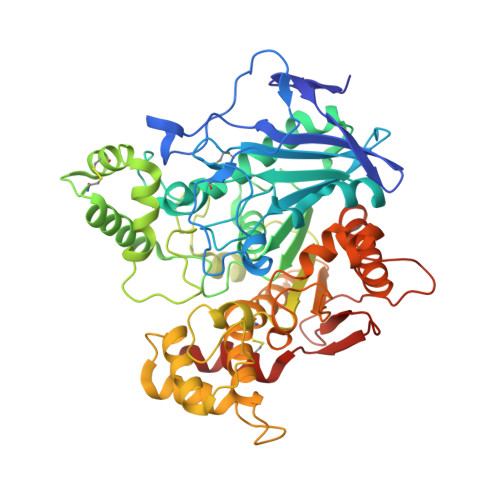
Human butyrylcholinesterase produced in insect cells: huprine-based affinity purification and crystal structure.
Brazzolotto, X., Wandhammer, M., Ronco, C., Trovaslet, M., Jean, L., Lockridge, O., Renard, P.Y., Nachon, F.(2012) FEBS J 279: 2905-2916
- PubMed: 22726956
- DOI: https://doi.org/10.1111/j.1742-4658.2012.08672.x
- Primary Citation of Related Structures:
4AQD - PubMed Abstract:
Butyrylcholinesterase (BChE) is a serine hydrolase that is present in all mammalian tissues. It can accommodate larger substrates or inhibitors than acetylcholinesterase (AChE), the enzyme responsible for hydrolysis of the neurotransmitter acetylcholine in the central nervous system and neuromuscular junctions. AChE is the specific target of organophosphorous pesticides and warfare nerve agents, and BChE is a stoichiometric bioscavenger. Conversion of BChE into a catalytic bioscavenger by rational design or designing reactivators specific to BChE required structural data obtained using a recombinant low-glycosylated human BChE expressed in Chinese hamster ovary cells. This expression system yields ≈ 1 mg of pure enzyme per litre of cell culture. Here, we report an improved expression system using insect cells with a fourfold higher yield for truncated human BChE with all glycosylation sites present. We developed a fast purification protocol for the recombinant protein using huprine-based affinity chromatography, which is superior to the classical procainamide-based affinity. The purified BChE crystallized under different conditions and space group than the recombinant low-glycosylated protein produced in Chinese hamster ovary cells. The crystals diffracted to 2.5 Å. The overall monomer structure is similar to the low-glycosylated structure except for the presence of the additional glycans. Remarkably, the carboxylic acid molecule systematically bound to the catalytic serine in the low-glycosylated structure is also present in this new structure, despite the different expression system, purification protocol and crystallization conditions.
Organizational Affiliation:
Département de Toxicologie, Institut de Recherche Biomédicale des Armées, La Tronche, France. xavier@brazzolotto.net