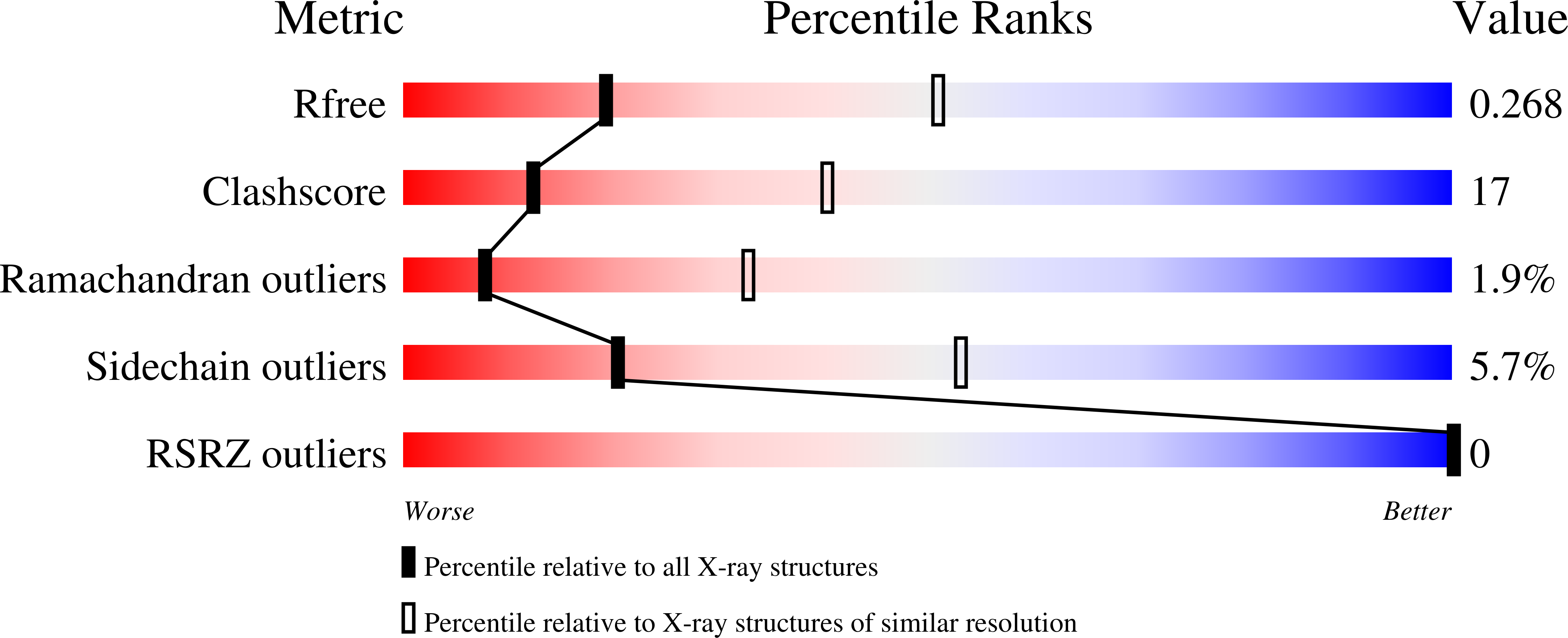
Mutational Analysis of Phenolic Acid Decarboxylase from Bacillus Subtilis (Bspad), which Converts Bio-Derived Phenolic Acids to Styrene Derivatives
Frank, A., Eborall, W., Hyde, R., Hart, S., Turkenburg, J.P., Grogan, G.(2012) Catal Sci Technol 2: 1568