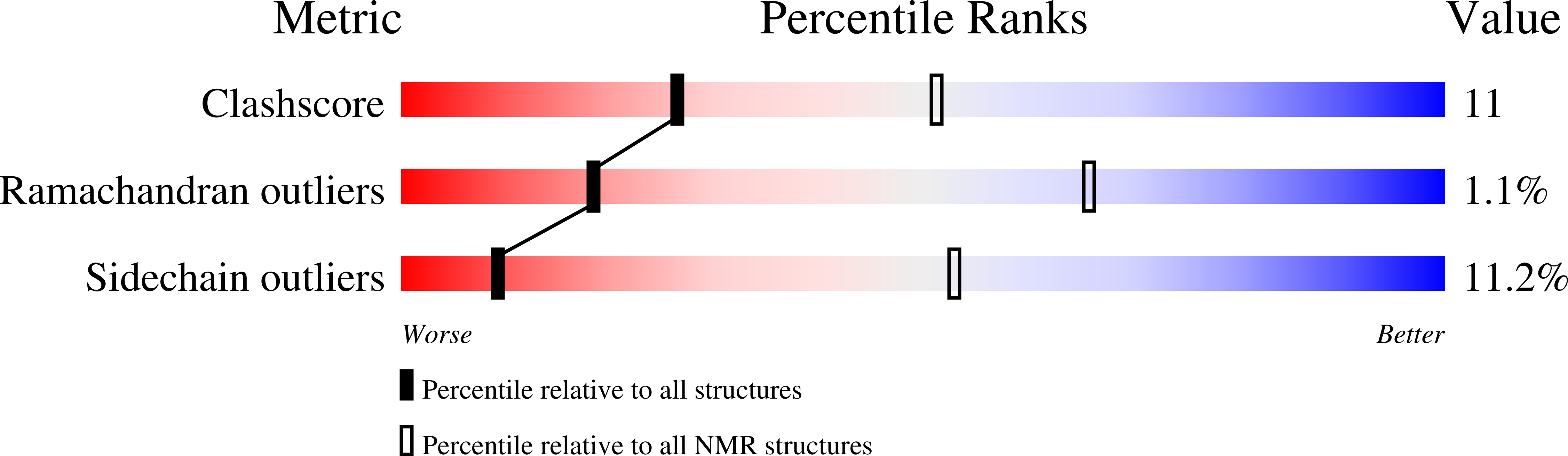Structural and Functional Analysis of the Deaf-1 and Bs69 Mynd Domains.
Kateb, F., Perrin, H., Tripsianes, K., Zou, P., Spadaccini, R., Bottomley, M., Franzmann, T.M., Buchner, J., Ansieau, S., Sattler, M.(2013) PLoS One 8: 54715
- PubMed: 23372760
- DOI: https://doi.org/10.1371/journal.pone.0054715
- Primary Citation of Related Structures:
4A24 - PubMed Abstract:
DEAF-1 is an important transcriptional regulator that is required for embryonic development and is linked to clinical depression and suicidal behavior in humans. It comprises various structural domains, including a SAND domain that mediates DNA binding and a MYND domain, a cysteine-rich module organized in a Cys(4)-Cys(2)-His-Cys (C4-C2HC) tandem zinc binding motif. DEAF-1 transcription regulation activity is mediated through interactions with cofactors such as NCoR and SMRT. Despite the important biological role of the DEAF-1 protein, little is known regarding the structure and binding properties of its MYND domain.Here, we report the solution structure, dynamics and ligand binding of the human DEAF-1 MYND domain encompassing residues 501-544 determined by NMR spectroscopy. The structure adopts a ββα fold that exhibits tandem zinc-binding sites with a cross-brace topology, similar to the MYND domains in AML1/ETO and other proteins. We show that the DEAF-1 MYND domain binds to peptides derived from SMRT and NCoR corepressors. The binding surface mapped by NMR titrations is similar to the one previously reported for AML1/ETO. The ligand binding and molecular functions of the related BS69 MYND domain were studied based on a homology model and mutational analysis. Interestingly, the interaction between BS69 and its binding partners (viral and cellular proteins) seems to require distinct charged residues flanking the predicted MYND domain fold, suggesting a different binding mode. Our findings demonstrate that the MYND domain is a conserved zinc binding fold that plays important roles in transcriptional regulation by mediating distinct molecular interactions with viral and cellular proteins.
Organizational Affiliation:
Institute of Structural Biology, Helmholtz Zentrum München, Neuherberg, Germany.