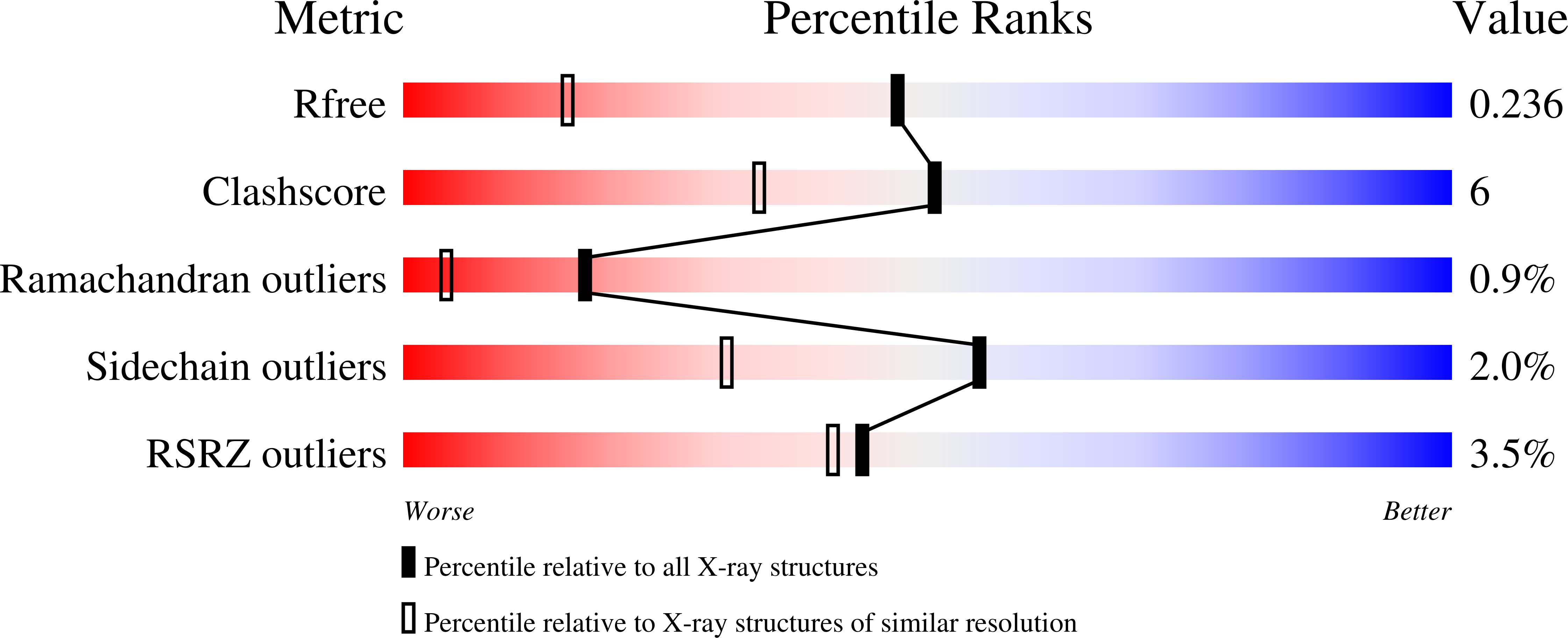
X-ray structure of p38 alpha bound to TAK-715: comparison with three classic inhibitors.
Azevedo, R., van Zeeland, M., Raaijmakers, H., Kazemier, B., de Vlieg, J., Oubrie, A.(2012) Acta Crystallogr D Biol Crystallogr 68: 1041-1050
- PubMed: 22868770
- DOI: https://doi.org/10.1107/S090744491201997X
- Primary Citation of Related Structures:
3ZS5, 3ZSG, 3ZSH, 3ZSI - PubMed Abstract:
The p38α mitogen-activated protein kinase regulates the synthesis of pro-inflammatory cytokines in response to stimulation by a diverse set of stress signals. Various different chemotypes and clinical candidates that inhibit p38α function have been reported over the years. In this publication, the novel structure of p38α cocrystallized with the clinical candidate TAK-715 is reported. Owing to the impact of crystallization conditions on the conformation of protein kinases (and in particular p38α), the structures of complexes of p38α with SB-203580, SCIO-469 and VX-745 have also been determined to enable in-depth comparison of ligand-induced protein conformations. The impact of experimental conditions on p38α-inhibitor complex structures, most importantly soaking versus cocrystallization, is discussed. Analysis of the structures and quantification of the protein-ligand interactions couples ligand-induced protein conformations to the number of interactions and to inhibitor selectivity against the human kinome. This shows that for the design of novel kinase inhibitors, selectivity is best obtained through maximization of the number of interactions throughout the ATP pocket and the exploitation of specific features in the active site.
Organizational Affiliation:
Merck Research Laboratories, MSD, PO Box 20, 5340 BH Oss, The Netherlands.