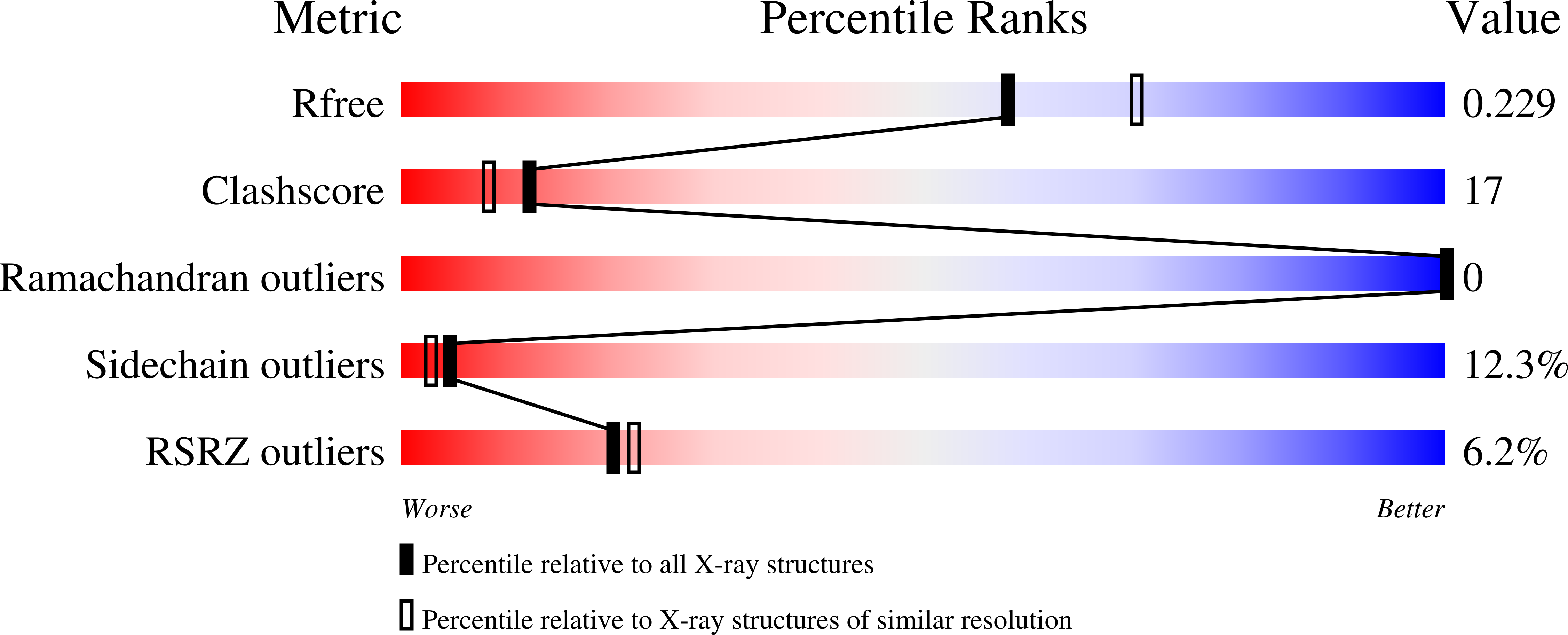
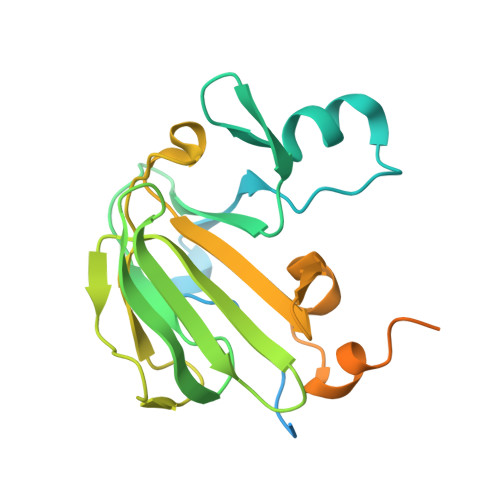
Structure of components of an intercellular channel complex in sporulating Bacillus subtilis.
Levdikov, V.M., Blagova, E.V., McFeat, A., Fogg, M.J., Wilson, K.S., Wilkinson, A.J.(2012) Proc Natl Acad Sci U S A 109: 5441-5445
- PubMed: 22431604
- DOI: https://doi.org/10.1073/pnas.1120087109
- Primary Citation of Related Structures:
3TUF - PubMed Abstract:
Following asymmetric cell division during spore formation in Bacillus subtilis, a forespore expressed membrane protein SpoIIQ, interacts across an intercellular space with a mother cell-expressed membrane protein, SpoIIIAH. Their interaction can serve as a molecular "ratchet" contributing to the migration of the mother cell membrane around that of the forespore in a phagocytosis-like process termed engulfment. Upon completion of engulfment, SpoIIQ and SpoIIIAH are integral components of a recently proposed intercellular channel allowing passage from the mother cell into the forespore of factors required for late gene expression in this compartment. Here we show that the extracellular domains of SpoIIQ and SpoIIIAH form a heterodimeric complex in solution. The crystal structure of this complex reveals that SpoIIQ has a LytM-like zinc-metalloprotease fold but with an incomplete zinc coordination sphere and no metal. SpoIIIAH has an α-helical subdomain and a protruding β-sheet subdomain, which mediates interactions with SpoIIQ. SpoIIIAH has sequence and structural homology to EscJ, a type III secretion system protein that forms a 24-fold symmetric ring. Superposition of the structures of SpoIIIAH and EscJ reveals that the SpoIIIAH protomer overlaps with two adjacent protomers of EscJ, allowing us to generate a dodecameric SpoIIIAH ring by using structural homology. Following this superposition, the SpoIIQ chains also form a closed dodecameric ring abutting the SpoIIIAH ring, producing an assembly surrounding a 60 Å channel. The dimensions and organization of the proposed complex suggest it is a plausible model for the extracellular component of a gap junction-like intercellular channel.
Organizational Affiliation:
Structural Biology Laboratory, Department of Chemistry, University of York, York YO10 5DD, United Kingdom.