Diversity in the C3b Convertase Contact Residues and Tertiary Structures of the Staphylococcal Complement Inhibitor (SCIN) Protein Family.
Garcia, B.L., Summers, B.J., Lin, Z., Ramyar, K.X., Ricklin, D., Kamath, D.V., Fu, Z.Q., Lambris, J.D., Geisbrecht, B.V.(2012) J Biol Chem 287: 628-640
- PubMed: 22086928
- DOI: https://doi.org/10.1074/jbc.M111.298984
- Primary Citation of Related Structures:
3T46, 3T47, 3T48, 3T49, 3T4A - PubMed Abstract:
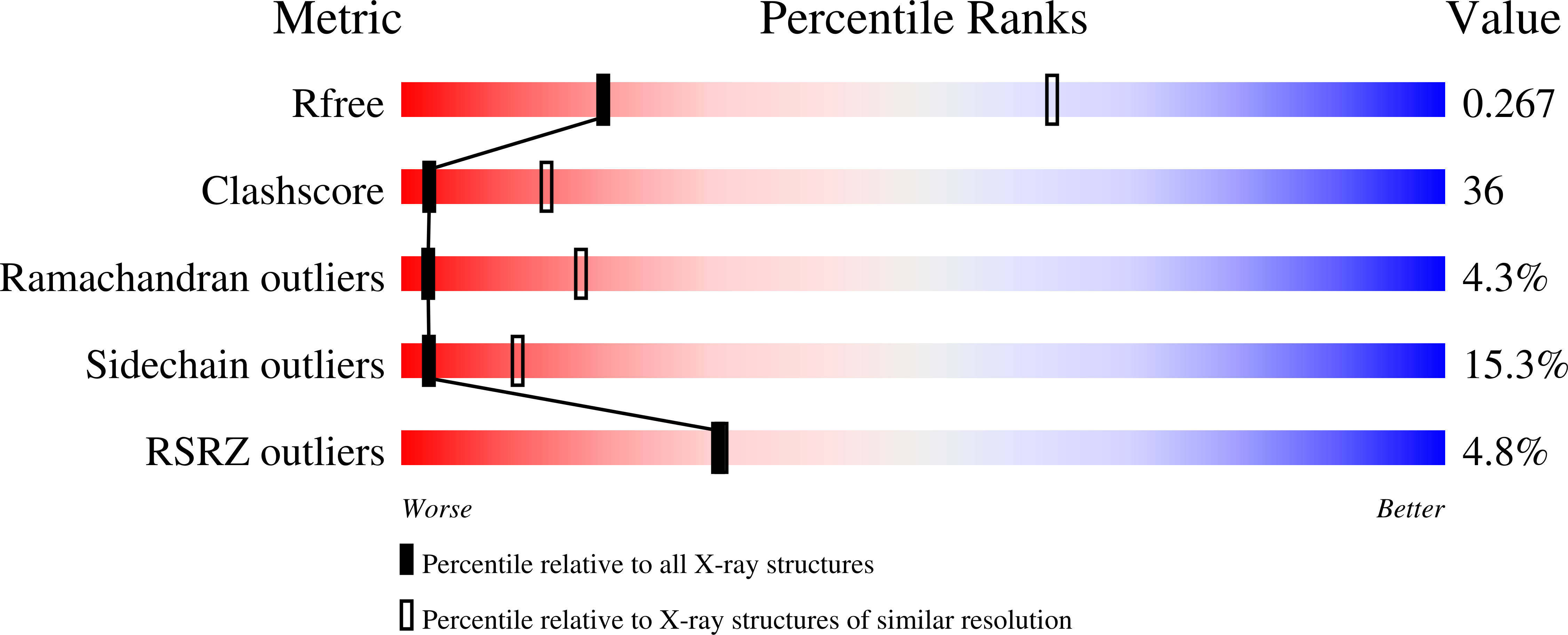
To survive in immune-competent hosts, the pathogen Staphylococcus aureus expresses and secretes a sophisticated array of proteins that inhibit the complement system. Among these are the staphylococcal complement inhibitors (SCIN), which are composed of three active proteins (SCIN-A, -B, and -C) and one purportedly inactive member (SCIN-D or ORF-D). Because previous work has focused almost exclusively on SCIN-A, we sought to provide initial structure/function information on additional SCIN proteins. To this end we determined crystal structures of an active, N-terminal truncation mutant of SCIN-B (denoted SCIN-B18-85) both free and bound to the C3c fragment of complement component C3 at 1.5 and 3.4 Å resolution, respectively. Comparison of the C3c/SCIN-B18-85 structure with that of C3c/SCIN-A revealed that both proteins target the same functional hotspot on the C3b/C3c surface yet harbor diversity in both the type of residues and interactions formed at their C3b/C3c interfaces. Most importantly, these structures allowed identification of Arg44 and Tyr51 as residues key for SCIN-B binding to C3b and subsequent inhibition of the AP C3 convertase. In addition, we also solved several crystal structures of SCIN-D to 1.3 Å limiting resolution. This revealed an unexpected structural deviation in the N-terminal α helix relative to SCIN-A and SCIN-B. Comparative analysis of both electrostatic potentials and surface complementarity suggest a physical explanation for the inability of SCIN-D to bind C3b/C3c. Together, these studies provide a more thorough understanding of immune evasion by S. aureus and enhance potential use of SCIN proteins as templates for design of complement targeted therapeutics.
Organizational Affiliation:
Division of Cell Biology and Biophysics, School of Biological Sciences, University of Missouri-Kansas City, Kansas City, Missouri 64110.