Crystal structure of human glycogen synthase kinase 3 beta (GSK3b) in complex with inhibitor 142
Cheng, R.K.Y., Rowan, F., Barker, J.J.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Glycogen synthase kinase-3 beta | 430 | Homo sapiens | Mutation(s): 0 Gene Names: GSK3B EC: 2.7.11.26 | 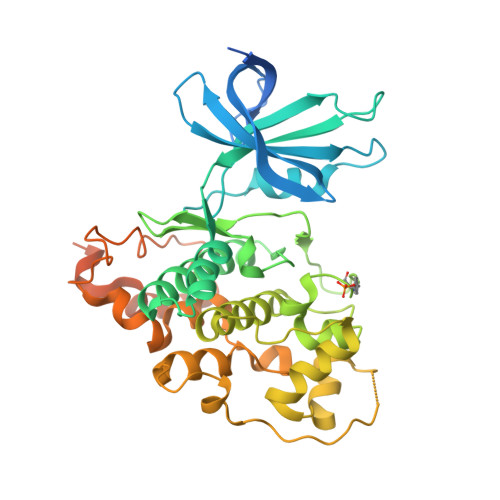 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P49841 (Homo sapiens) Explore P49841 Go to UniProtKB: P49841 | |||||
PHAROS: P49841 GTEx: ENSG00000082701 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P49841 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| OFT Query on OFT | C [auth A], F [auth B] | (3Z)-N,N-diethyl-3-[(3E)-3-(hydroxyimino)-1,3-dihydro-2H-indol-2-ylidene]-2-oxo-2,3-dihydro-1H-indole-5-sulfonamide C20 H20 N4 O4 S ZZYLNEHSVPINPK-LCLDVOFVSA-N |  | ||
| MPD Query on MPD | H [auth B] | (4S)-2-METHYL-2,4-PENTANEDIOL C6 H14 O2 SVTBMSDMJJWYQN-YFKPBYRVSA-N |  | ||
| MLA Query on MLA | D [auth A], G [auth B] | MALONIC ACID C3 H4 O4 OFOBLEOULBTSOW-UHFFFAOYSA-N |  | ||
| FMT Query on FMT | E [auth A] | FORMIC ACID C H2 O2 BDAGIHXWWSANSR-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| PTR Query on PTR | A, B | L-PEPTIDE LINKING | C9 H12 N O6 P |  | TYR |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 67.52 | α = 90 |
| b = 110.78 | β = 97.08 |
| c = 67.91 | γ = 90 |
| Software Name | Purpose |
|---|---|
| GDA | data collection |
| PHASER | phasing |
| REFMAC | refinement |
| XDS | data reduction |
| SCALA | data scaling |