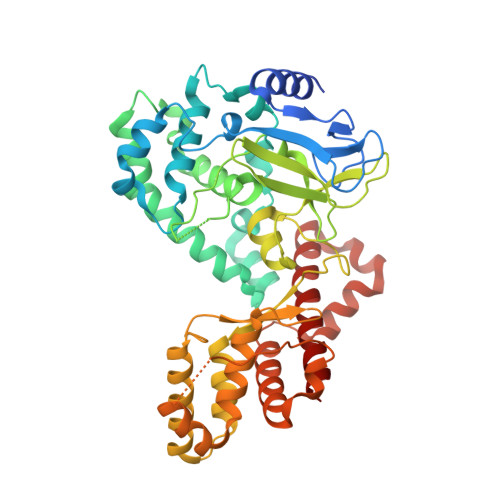
Structural basis of SETD6-mediated regulation of the NF-kB network via methyl-lysine signaling.
Chang, Y., Levy, D., Horton, J.R., Peng, J., Zhang, X., Gozani, O., Cheng, X.(2011) Nucleic Acids Res 39: 6380-6389
- PubMed: 21515635
- DOI: https://doi.org/10.1093/nar/gkr256
- Primary Citation of Related Structures:
3QXY, 3RC0 - PubMed Abstract:
SET domain containing 6 (SETD6) monomethylates the RelA subunit of nuclear factor kappa B (NF-κB). The ankyrin repeats of G9a-like protein (GLP) recognizes RelA monomethylated at Lys310. Adjacent to Lys310 is Ser311, a known phosphorylation site of RelA. Ser311 phosphorylation inhibits Lys310 methylation by SETD6 as well as binding of Lys310me1 by GLP. The structure of SETD6 in complex with RelA peptide containing the methylation site, in the presence of S-adenosyl-L-methionine, reveals a V-like protein structure and suggests a model for NF-κB binding to SETD6. In addition, structural modeling of the GLP ankyrin repeats bound to Lys310me1 peptide provides insight into the molecular basis for inhibition of Lys310me1 binding by Ser311 phosphorylation. Together, these findings provide a structural explanation for a key cellular signaling pathway centered on RelA Lys310 methylation, which is generated by SETD6 and recognized by GLP, and incorporate a methylation-phosphorylation switch of adjacent lysine and serine residues. Finally, SETD6 is structurally similar to the Rubisco large subunit methyltransferase. Given the restriction of Rubisco to plant species, this particular appearance of the protein lysine methyltransferase has been evolutionarily well conserved.
Organizational Affiliation:
Department of Biochemistry, Emory University School of Medicine, 1510 Clifton Road, Atlanta, GA 30322, USA.