Structure of p300 bound to MEF2 on DNA reveals a mechanism of enhanceosome assembly.
He, J., Ye, J., Cai, Y., Riquelme, C., Liu, J.O., Liu, X., Han, A., Chen, L.(2011) Nucleic Acids Res 39: 4464-4474
- PubMed: 21278418
- DOI: https://doi.org/10.1093/nar/gkr030
- Primary Citation of Related Structures:
3P57 - PubMed Abstract:
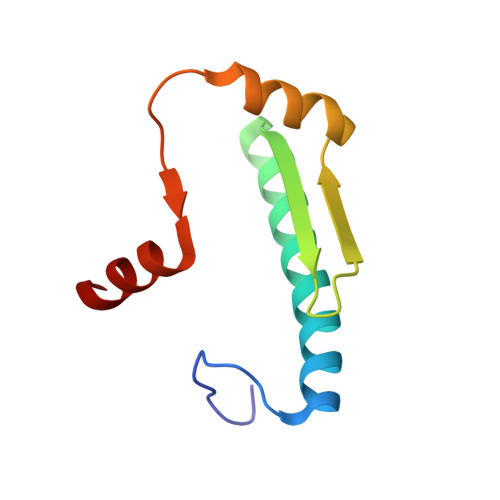
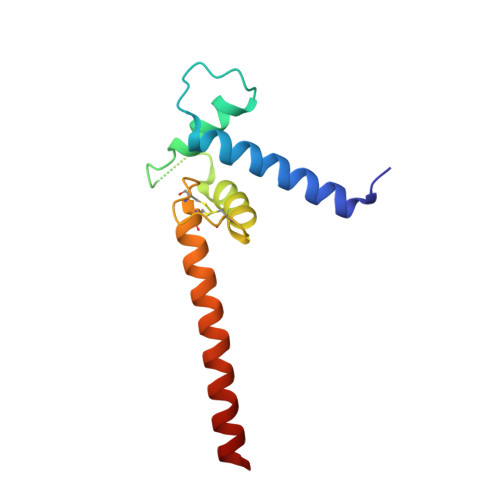
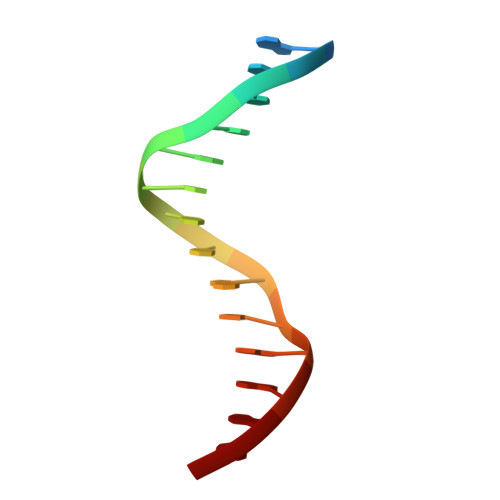
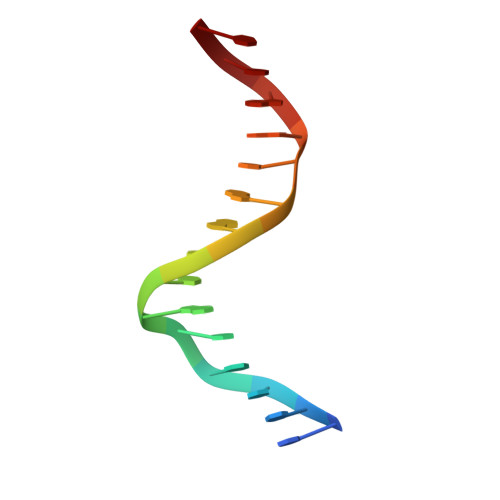
Transcription co-activators CBP and p300 are recruited by sequence-specific transcription factors to specific genomic loci to control gene expression. A highly conserved domain in CBP/p300, the TAZ2 domain, mediates direct interaction with a variety of transcription factors including the myocyte enhancer factor 2 (MEF2). Here we report the crystal structure of a ternary complex of the p300 TAZ2 domain bound to MEF2 on DNA at 2.2Å resolution. The structure reveals three MEF2:DNA complexes binding to different sites of the TAZ2 domain. Using structure-guided mutations and a mammalian two-hybrid assay, we show that all three interfaces contribute to the binding of MEF2 to p300, suggesting that p300 may use one of the three interfaces to interact with MEF2 in different cellular contexts and that one p300 can bind three MEF2:DNA complexes simultaneously. These studies, together with previously characterized TAZ2 complexes bound to different transcription factors, demonstrate the potency and versatility of TAZ2 in protein-protein interactions. Our results also support a model wherein p300 promotes the assembly of a higher-order enhanceosome by simultaneous interactions with multiple DNA-bound transcription factors.
Organizational Affiliation:
MOE Key Laboratory for Cell Biology, School of Life Sciences, Xiamen University, Xiamen, Fujian 361005, China.