NADH oxidase activity of Bacillus subtilis nitroreductase NfrA1: insight into its biological role.
Cortial, S., Chaignon, P., Iorga, B.I., Aymerich, S., Truan, G., Gueguen-Chaignon, V., Meyer, P., Morera, S., Ouazzani, J.(2010) FEBS Lett 584: 3916-3922
- PubMed: 20727352
- DOI: https://doi.org/10.1016/j.febslet.2010.08.019
- Primary Citation of Related Structures:
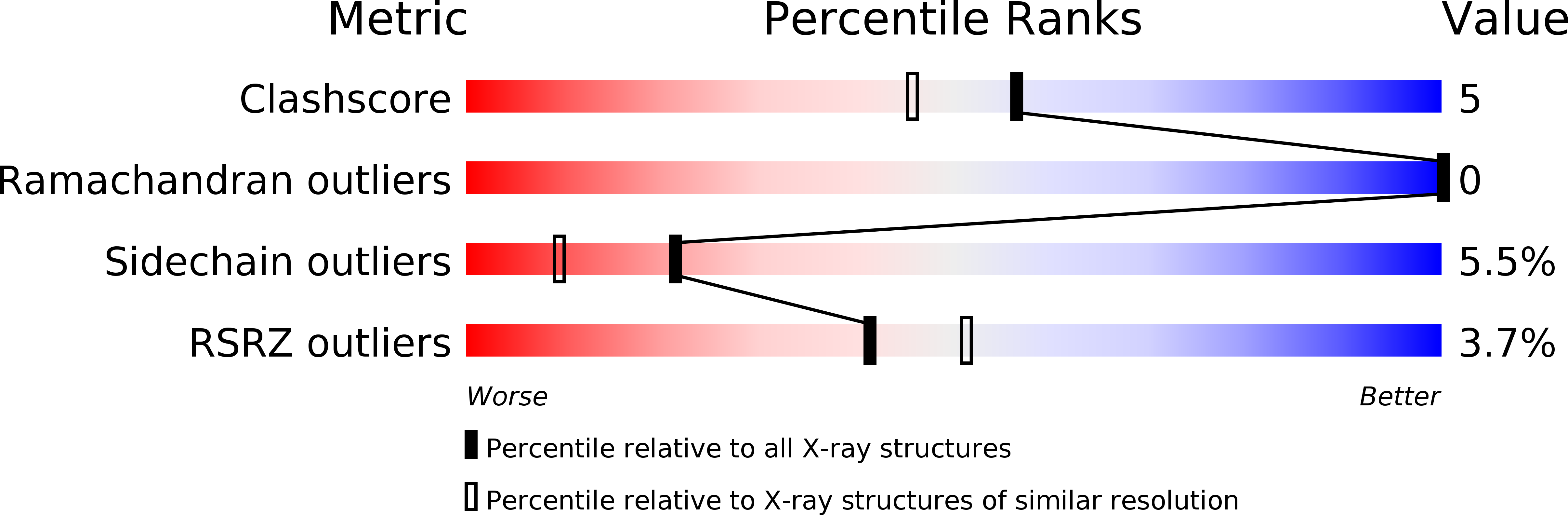
3N2S - PubMed Abstract:
NfrA1 nitroreductase from the Gram-positive bacterium Bacillus subtilis is a member of the NAD(P)H/FMN oxidoreductase family. Here, we investigated the reactivity, the structure and kinetics of NfrA1, which could provide insight into the unclear biological role of this enzyme. We could show that NfrA1 possesses an NADH oxidase activity that leads to high concentrations of oxygen peroxide and an NAD(+) degrading activity leading to free nicotinamide. Finally, we showed that NfrA1 is able to rapidly scavenge H(2)O(2) produced during the oxidative process or added exogenously.
Organizational Affiliation:
Institut de Chimie des Substances Naturelles (ICSN), CNRS UPR 2301, Gif-sur-Yvette, France.