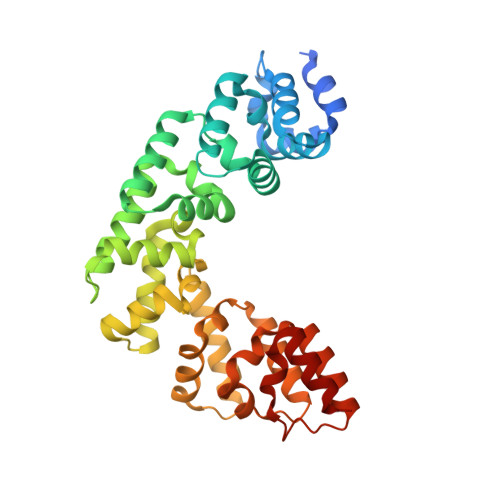
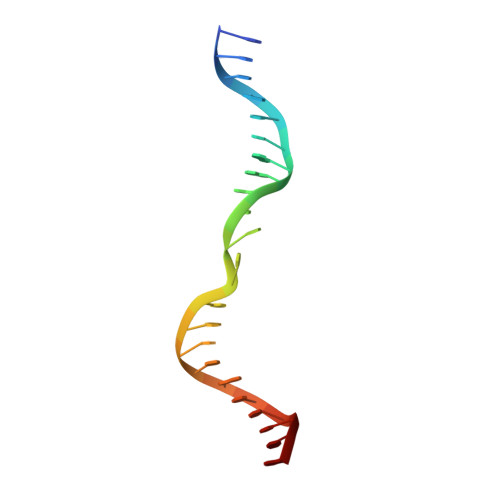
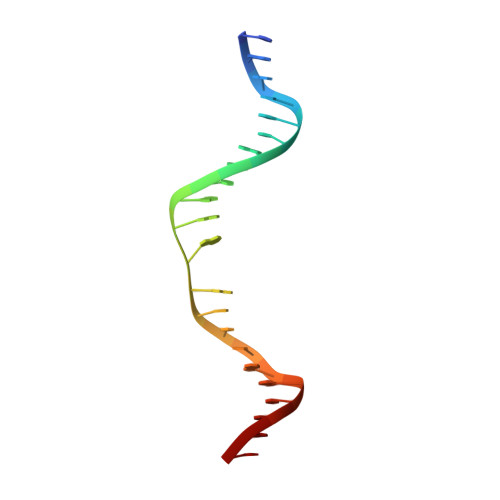
Helix unwinding and base flipping enable human MTERF1 to terminate mitochondrial transcription.
Yakubovskaya, E., Mejia, E., Byrnes, J., Hambardjieva, E., Garcia-Diaz, M.(2010) Cell 141: 982-993
- PubMed: 20550934
- DOI: https://doi.org/10.1016/j.cell.2010.05.018
- Primary Citation of Related Structures:
3MVA, 3MVB - PubMed Abstract:
Defects in mitochondrial gene expression are associated with aging and disease. Mterf proteins have been implicated in modulating transcription, replication and protein synthesis. We have solved the structure of a member of this family, the human mitochondrial transcriptional terminator MTERF1, bound to dsDNA containing the termination sequence. The structure indicates that upon sequence recognition MTERF1 unwinds the DNA molecule, promoting eversion of three nucleotides. Base flipping is critical for stable binding and transcriptional termination. Additional structural and biochemical results provide insight into the DNA binding mechanism and explain how MTERF1 recognizes its target sequence. Finally, we have demonstrated that the mitochondrial pathogenic G3249A and G3244A mutations interfere with key interactions for sequence recognition, eliminating termination. Our results provide insight into the role of mterf proteins and suggest a link between mitochondrial disease and the regulation of mitochondrial transcription.
Organizational Affiliation:
Department of Pharmacological Sciences. Stony Brook University, Stony Brook, NY 11794, USA.