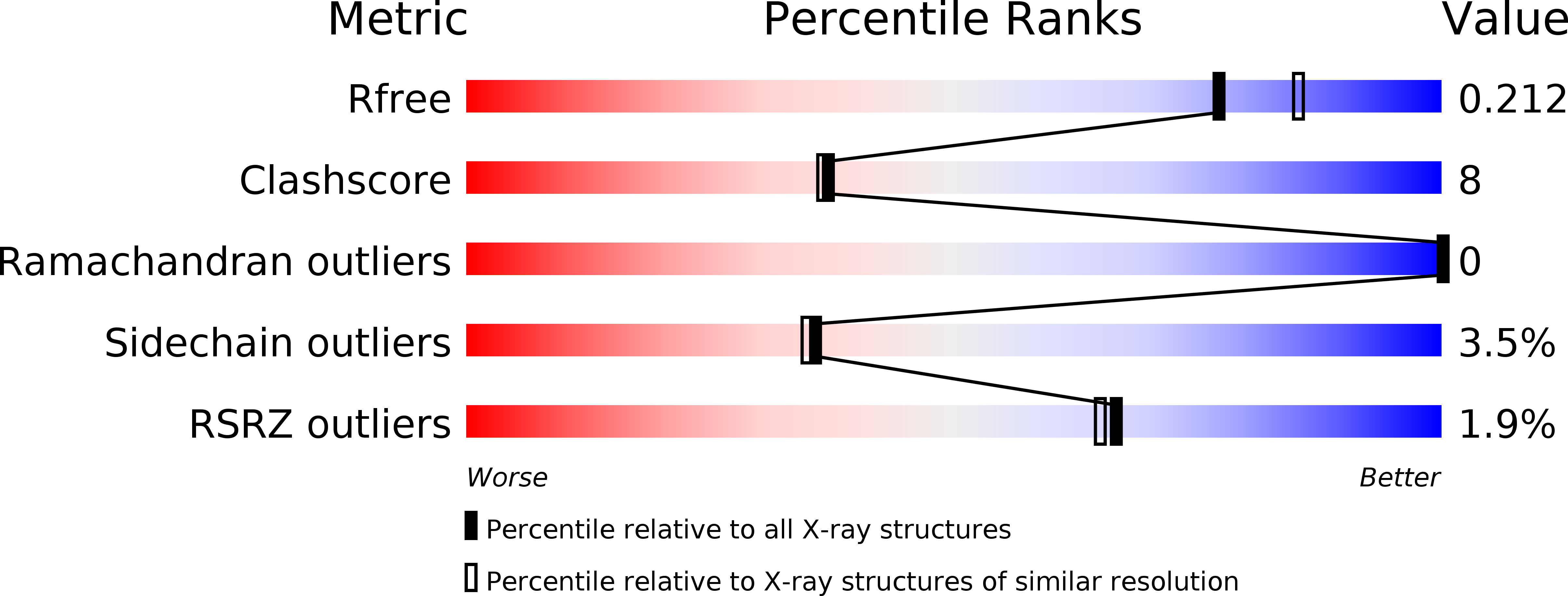
The protein tyrosine phosphatases PTPRZ and PTPRG bind to distinct members of the contactin family of neural recognition molecules.
Bouyain, S., Watkins, D.J.(2010) Proc Natl Acad Sci U S A 107: 2443-2448
- PubMed: 20133774
- DOI: https://doi.org/10.1073/pnas.0911235107
- Primary Citation of Related Structures:
3JXA, 3JXF, 3JXG, 3JXH, 3KLD - PubMed Abstract:
The receptor protein tyrosine phosphatases gamma (PTPRG) and zeta (PTPRZ) are expressed primarily in the nervous system and mediate cell adhesion and signaling events during development. We report here the crystal structures of the carbonic anhydrase-like domains of PTPRZ and PTPRG and show that these domains interact directly with the second and third immunoglobulin repeats of the members of the contactin (CNTN) family of neural recognition molecules. Interestingly, these receptors exhibit distinct specificities: PTPRZ binds only to CNTN1, whereas PTPRG interacts with CNTN3, 4, 5, and 6. Furthermore, we present crystal structures of the four N-terminal immunoglobulin repeats of mouse CNTN4 both alone and in complex with the carbonic anhydrase-like domain of mouse PTPRG. In these structures, the N-terminal region of CNTN4 adopts a horseshoe-like conformation found also in CNTN2 and most likely in all CNTNs. This restrained conformation of the second and third immunoglobulin domains creates a binding site that is conserved among CNTN3, 4, 5, and 6. This site contacts a discrete region of PTPRG composed primarily of an extended beta-hairpin loop found in both PTPRG and PTPRZ. Overall, these findings implicate PTPRG, PTPRZ and CNTNs as a group of receptors and ligands involved in the manifold recognition events that underlie the construction of neural networks.
Organizational Affiliation:
Division of Molecular Biology and Biochemistry, School of Biological Sciences, University of Missouri-Kansas City, Kansas City, MO 64110, USA. bouyains@umkc.edu