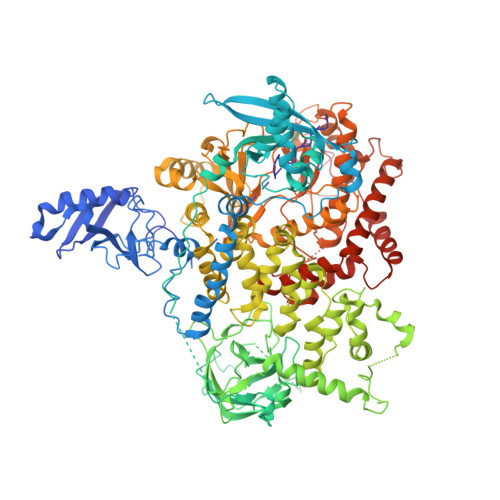
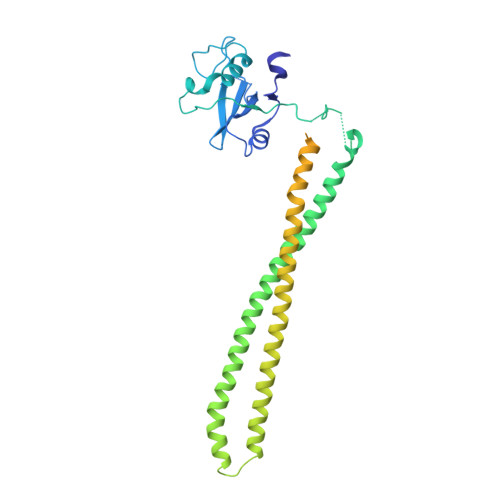
A frequent kinase domain mutation that changes the interaction between PI3K{alpha} and the membrane.
Mandelker, D., Gabelli, S.B., Schmidt-Kittler, O., Zhu, J., Cheong, I., Huang, C.H., Kinzler, K.W., Vogelstein, B., Amzel, L.M.(2009) Proc Natl Acad Sci U S A 106: 16996-17001
- PubMed: 19805105
- DOI: https://doi.org/10.1073/pnas.0908444106
- Primary Citation of Related Structures:
3HHM, 3HIZ - PubMed Abstract:
Mutations in oncogenes often promote tumorigenesis by changing the conformation of the encoded proteins, thereby altering enzymatic activity. The PIK3CA oncogene, which encodes p110alpha, the catalytic subunit of phosphatidylinositol 3-kinase alpha (PI3Kalpha), is one of the two most frequently mutated oncogenes in human cancers. We report the structure of the most common mutant of p110alpha in complex with two interacting domains of its regulatory partner (p85alpha), both free and bound to an inhibitor (wortmannin). The N-terminal SH2 (nSH2) domain of p85alpha is shown to form a scaffold for the entire enzyme complex, strategically positioned to communicate extrinsic signals from phosphopeptides to three distinct regions of p110alpha. Moreover, we found that Arg-1047 points toward the cell membrane, perpendicular to the orientation of His-1047 in the WT enzyme. Surprisingly, two loops of the kinase domain that contact the cell membrane shift conformation in the oncogenic mutant. Biochemical assays revealed that the enzymatic activity of the p110alpha His1047Arg mutant is differentially regulated by lipid membrane composition. These structural and biochemical data suggest a previously undescribed mechanism for mutational activation of a kinase that involves perturbation of its interaction with the cellular membrane.
Organizational Affiliation:
The Ludwig Center for Cancer Genetics and Therapeutics and Howard Hughes Medical Institute, Johns Hopkins Kimmel Cancer Center, Baltimore, MD 21231, USA.