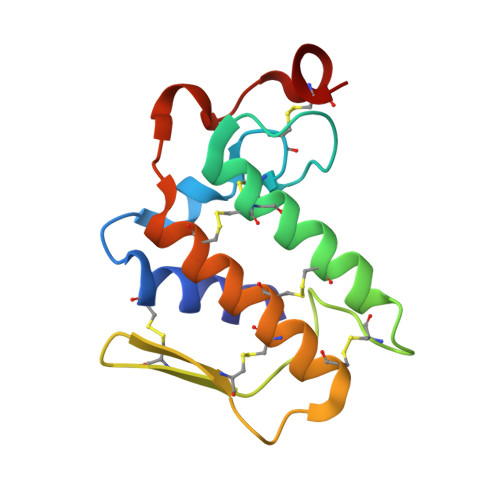
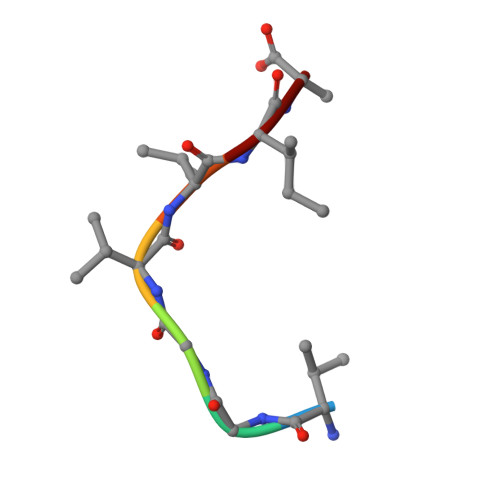
Crystal Structure of the Complex Formed Between a New Isoform of Phospholipase A2 with C-terminal Amyloid Beta Heptapeptide at 2 A Resolution
Mirza, Z., Vikram, G., Singh, N., Sinha, M., Bhushan, A., Sharma, S., Srinivasan, A., Kaur, P., Singh, T.P.To be published.