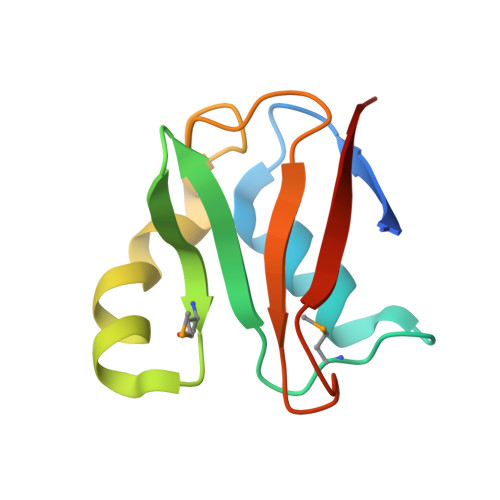
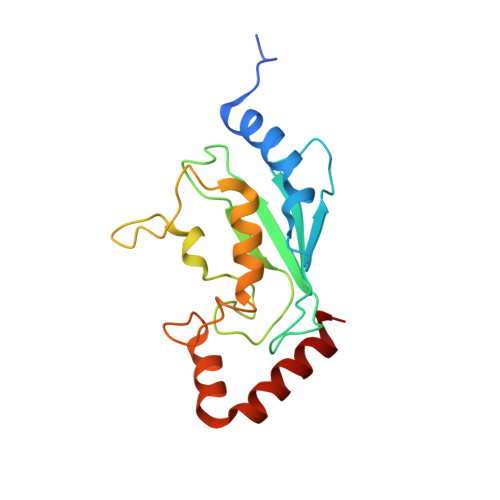
E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification
Huang, D.T., Ayrault, O., Hunt, H.W., Taherbhoy, A.M., Duda, D.M., Scott, D.C., Borg, L.A., Neale, G., Murray, P.J., Roussel, M.F., Schulman, B.A.(2009) Mol Cell 33: 483-495
- PubMed: 19250909
- DOI: https://doi.org/10.1016/j.molcel.2009.01.011
- Primary Citation of Related Structures:
3FN1 - PubMed Abstract:
Ubiquitin and ubiquitin-like proteins (UBLs) are directed to targets by cascades of E1, E2, and E3 enzymes. The largest ubiquitin E3 subclass consists of cullin-RING ligases (CRLs), which contain one each of several cullins (CUL1, -2, -3, -4, or -5) and RING proteins (RBX1 or -2). CRLs are activated by ligation of the UBL NEDD8 to a conserved cullin lysine. How is cullin NEDD8ylation specificity established? Here we report that, like UBE2M (also known as UBC12), the previously uncharacterized E2 UBE2F is a NEDD8-conjugating enzyme in vitro and in vivo. Biochemical and structural analyses indicate how plasticity of hydrophobic E1-E2 interactions and E1 conformational flexibility allow one E1 to charge multiple E2s. The E2s have distinct functions, with UBE2M/RBX1 and UBE2F/RBX2 displaying different target cullin specificities. Together, these studies reveal the molecular basis for and functional importance of hierarchical expansion of the NEDD8 conjugation system in establishing selective CRL activation.
Organizational Affiliation:
Department of Structural Biology, St. Jude Children's Research Hospital, Memphis, TN 38105, USA.