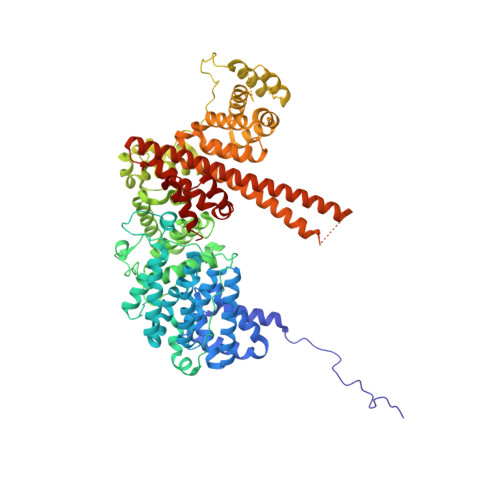
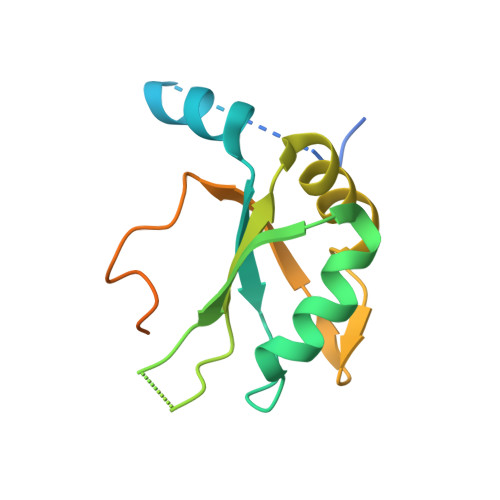
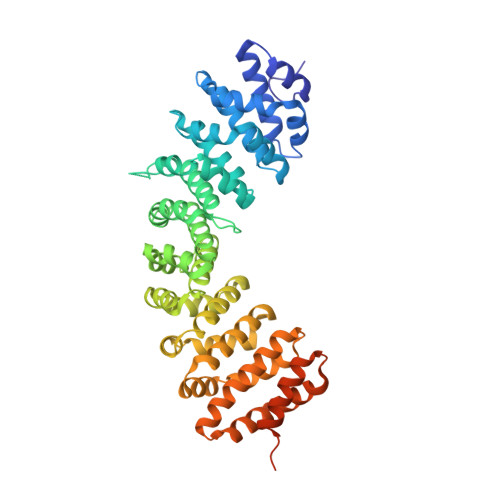
The molecular basis for the regulation of the cap-binding complex by the importins.
Dias, S.M., Wilson, K.F., Rojas, K.S., Ambrosio, A.L., Cerione, R.A.(2009) Nat Struct Mol Biol 16: 930-937
- PubMed: 19668212
- DOI: https://doi.org/10.1038/nsmb.1649
- Primary Citation of Related Structures:
3FEX, 3FEY - PubMed Abstract:
The binding of capped RNAs to the cap-binding complex (CBC) in the nucleus, and their dissociation from the CBC in the cytosol, represent essential steps in RNA processing. Here we show how the nucleocytoplasmic transport proteins importin-alpha and importin-beta have key roles in regulating these events. As a first step toward understanding the molecular basis for this regulation, we determined a 2.2-A resolution X-ray structure for a CBC-importin-alpha complex that provides a detailed picture for how importin-alpha binds to the CBP80 subunit of the CBC. Through a combination of biochemical studies, X-ray crystallographic information and small-angle scattering experiments, we then determined how importin-beta binds to the CBC through its CBP20 subunit. Together, these studies enable us to propose a model describing how importin-beta stimulates the dissociation of capped RNA from the CBC in the cytosol following its nuclear export.
Organizational Affiliation:
Department of Molecular Medicine, College of Veterinary Medicine, Cornell University, Ithaca, New York, USA.