Crystal structures and evolutionary relationship of two different lipoamide dehydrogenase(E3s) from Thermus thermophilus
Kondo, H., Hossain, M.T., Adachi, W., Nakai, T., Kamiya, N., Kuramitsu, S., Sekiguchi, T., Takenaka, A.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 2-oxoglutarate dehydrogenase E3 component | 455 | Thermus thermophilus HB8 | Mutation(s): 0 EC: 1.8.1.4 | 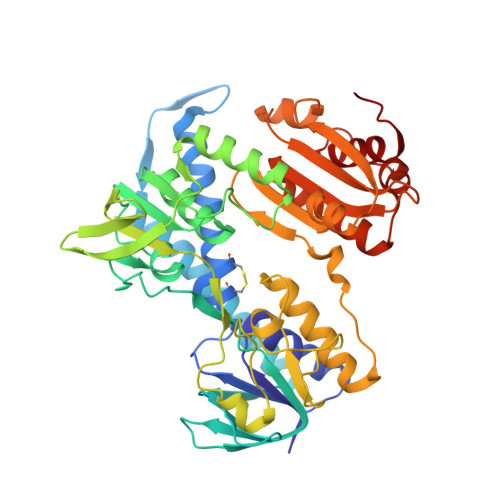 | |
UniProt | |||||
Find proteins for Q5SLK6 (Thermus thermophilus (strain ATCC 27634 / DSM 579 / HB8)) Explore Q5SLK6 Go to UniProtKB: Q5SLK6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5SLK6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FAD Query on FAD | D [auth A], H [auth B] | FLAVIN-ADENINE DINUCLEOTIDE C27 H33 N9 O15 P2 VWWQXMAJTJZDQX-UYBVJOGSSA-N |  | ||
| PO4 Query on PO4 | G [auth B] | PHOSPHATE ION O4 P NBIIXXVUZAFLBC-UHFFFAOYSA-K |  | ||
| CO3 Query on CO3 | C [auth A], E [auth B], F [auth B] | CARBONATE ION C O3 BVKZGUZCCUSVTD-UHFFFAOYSA-L |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 54.411 | α = 90 |
| b = 105.76 | β = 92.65 |
| c = 84.907 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| HKL-2000 | data collection |
| DENZO | data reduction |
| SCALEPACK | data scaling |
| AMoRE | phasing |