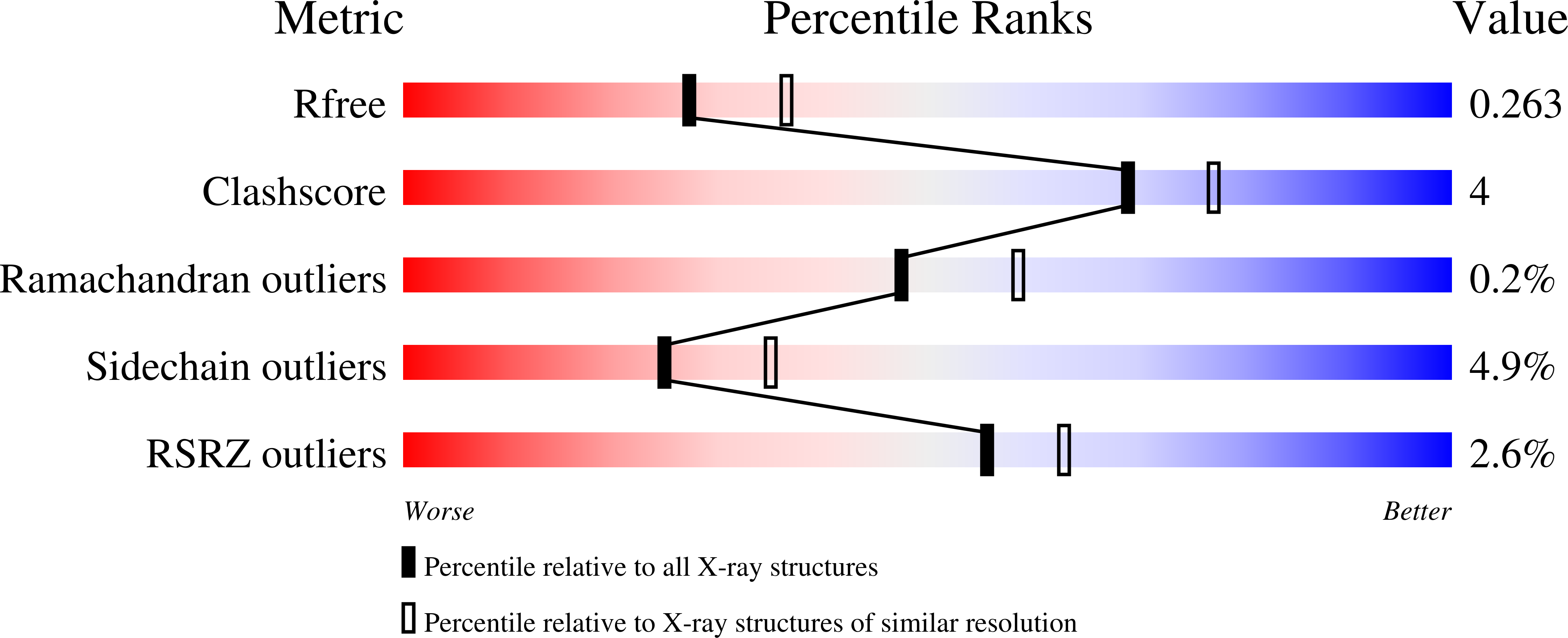
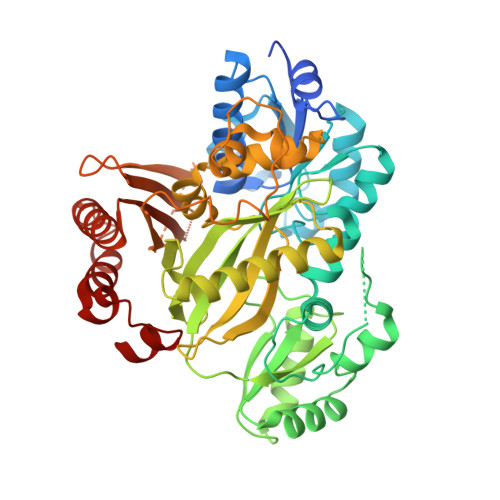
Crystal Structure of Human Acetyl-Coa Carboxylase 1, Biotin Carboxylase (Bc) Domain
Muniz, J.R.C., Froese, D.S., Krysztofinska, E., Vollmar, M., Beltrami, A., Krojer, T., Allerston, C.K., von Delft, F., Arrowsmith, C.H., Edwards, A.M., Weigelt, J., Bountra, C., Yue, W.W., Oppermann, U.To be published.