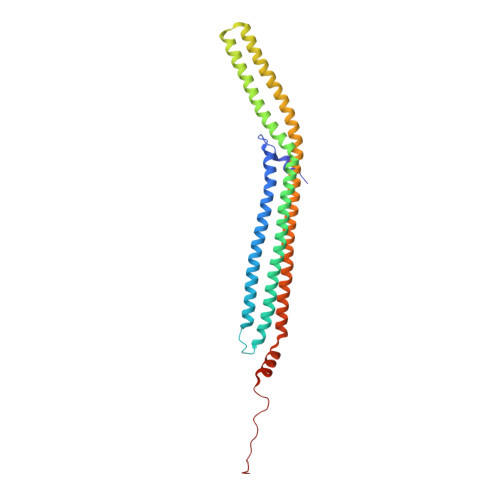
Structure and Analysis of Fcho2 F-Bar Domain: A Dimerizing and Membrane Recruitment Module that Effects Membrane Curvature.
Henne, W.M., Kent, H.M., Ford, M.J.G., Hedge, B.G., Daumke, O., Butler, P.J., Mittal, R., Langen, R., Evans, P.R., Mcmahon, H.T.(2007) Structure 15: 839
- PubMed: 17540576
- DOI: https://doi.org/10.1016/j.str.2007.05.002
- Primary Citation of Related Structures:
2V0O - PubMed Abstract:
A spectrum of membrane curvatures exists within cells, and proteins have evolved different modules to detect, create, and maintain these curvatures. Here we present the crystal structure of one such module found within human FCHo2. This F-BAR (extended FCH) module consists of two F-BAR domains, forming an intrinsically curved all-helical antiparallel dimer with a Kd of 2.5 microM. The module binds liposomes via a concave face, deforming them into tubules with variable diameters of up to 130 nm. Pulse EPR studies showed the membrane-bound dimer is the same as the crystal dimer, although the N-terminal helix changed conformation on membrane binding. Mutation of a phenylalanine on this helix partially attenuated narrow tubule formation, and resulted in a gain of curvature sensitivity. This structure shows a distant relationship to curvature-sensing BAR modules, and suggests how similar coiled-coil architectures in the BAR superfamily have evolved to expand the repertoire of membrane-sculpting possibilities.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Hills Road, Cambridge, CB1 0QH, United Kingdom.