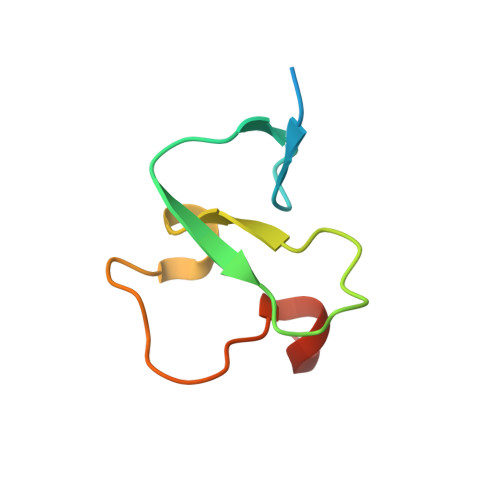
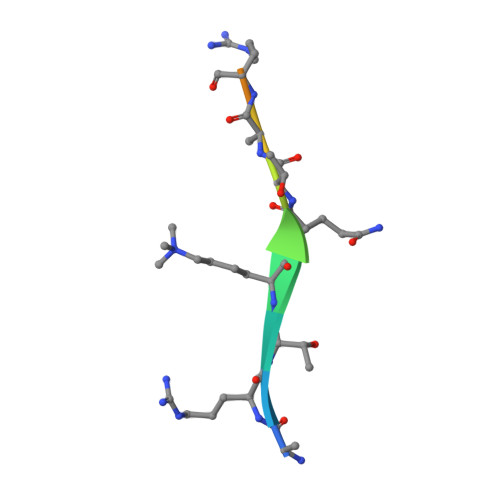
Histone H3K4me3 binding is required for the DNA repair and apoptotic activities of ING1 tumor suppressor.
Pena, P.V., Hom, R.A., Hung, T., Lin, H., Kuo, A.J., Wong, R.P., Subach, O.M., Champagne, K.S., Zhao, R., Verkhusha, V.V., Li, G., Gozani, O., Kutateladze, T.G.(2008) J Mol Biol 380: 303-312
- PubMed: 18533182
- DOI: https://doi.org/10.1016/j.jmb.2008.04.061
- Primary Citation of Related Structures:
2QIC - PubMed Abstract:
Inhibitor of growth 1 (ING1) is implicated in oncogenesis, DNA damage repair, and apoptosis. Mutations within the ING1 gene and altered expression levels of ING1 are found in multiple human cancers. Here, we show that both DNA repair and apoptotic activities of ING1 require the interaction of the C-terminal plant homeodomain (PHD) finger with histone H3 trimethylated at Lys4 (H3K4me3). The ING1 PHD finger recognizes methylated H3K4 but not other histone modifications as revealed by the peptide microarrays. The molecular mechanism of the histone recognition is elucidated based on a 2.1 A-resolution crystal structure of the PHD-H3K4me3 complex. The K4me3 occupies a deep hydrophobic pocket formed by the conserved Y212 and W235 residues that make cation-pi contacts with the trimethylammonium group. Both aromatic residues are essential in the H3K4me3 recognition, as substitution of these residues with Ala disrupts the interaction. Unlike the wild-type ING1, the W235A mutant, overexpressed in the stable clones of melanoma cells or in HT1080 cells, was unable to stimulate DNA repair after UV irradiation or promote DNA-damage-induced apoptosis, indicating that H3K4me3 binding is necessary for these biological functions of ING1. Furthermore, N216S, V218I, and G221V mutations, found in human malignancies, impair the ability of ING1 to associate with H3K4me3 or to induce nucleotide repair and cell death, linking the tumorigenic activity of ING1 with epigenetic regulation. Together, our findings reveal the critical role of the H3K4me3 interaction in mediating cellular responses to genotoxic stresses and offer new insight into the molecular mechanism underlying the tumor suppressive activity of ING1.
Organizational Affiliation:
Department of Pharmacology, University of Colorado Health Sciences Center, 12801 East 17th Avenue, Aurora, CO 80045-0511, USA.