The crystal structure of the human RAC3 in complex with the CRIB domain of human p21-activated kinase 4 (PAK4)
Ugochukwu, E., Yang, X., Elkins, J.M., Burgess-Brown, N., Knapp, S., Doyle, D.A.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ras-related C3 botulinum toxin substrate 3 | 179 | Homo sapiens | Mutation(s): 0 Gene Names: RAC3 | 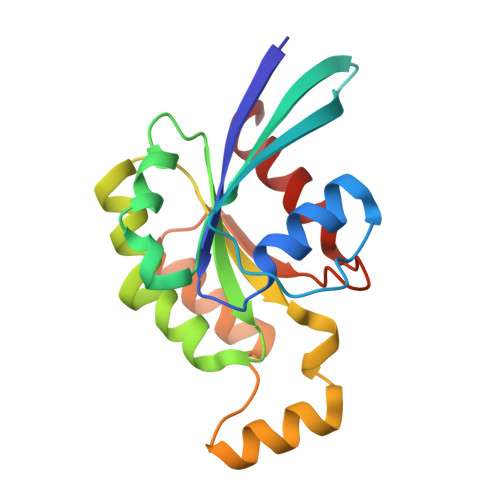 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P60763 (Homo sapiens) Explore P60763 Go to UniProtKB: P60763 | |||||
PHAROS: P60763 GTEx: ENSG00000169750 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P60763 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Serine/threonine-protein kinase PAK 4 | 35 | Homo sapiens | Mutation(s): 0 Gene Names: PAK4, KIAA1142 EC: 2.7.11.1 | 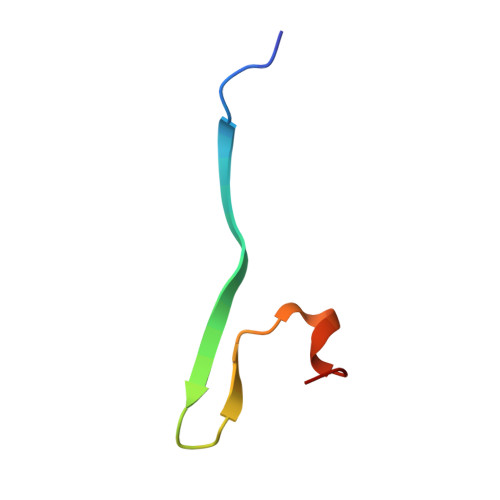 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O96013 (Homo sapiens) Explore O96013 Go to UniProtKB: O96013 | |||||
PHAROS: O96013 GTEx: ENSG00000130669 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O96013 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GCP Query on GCP | CA [auth C] FA [auth G] HA [auth D] LA [auth E] NA [auth H] | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER C11 H18 N5 O13 P3 PHBDHXOBFUBCJD-KQYNXXCUSA-N |  | ||
| EDO Query on EDO | DA [auth C], IA [auth D], W [auth F] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| CL Query on CL | BA [auth C] KA [auth E] OA [auth N] PA [auth O] QA [auth P] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| MG Query on MG | AA [auth C] EA [auth G] GA [auth D] JA [auth E] MA [auth H] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 50.843 | α = 88.58 |
| b = 73.859 | β = 87.76 |
| c = 133.84 | γ = 70.64 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| MAR345 | data collection |
| MOSFLM | data reduction |
| CCP4 | data scaling |
| PHASER | phasing |