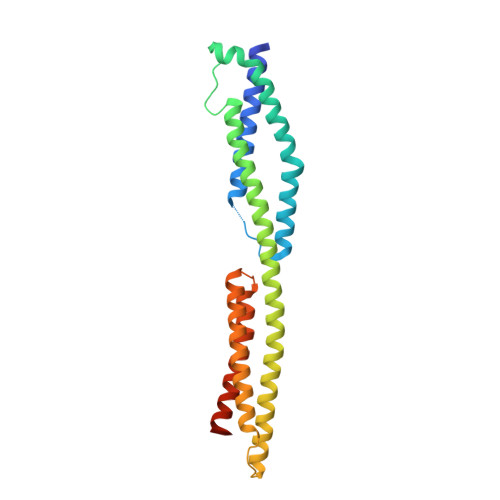
The structure of a tandem pair of spectrin repeats of plectin reveals a modular organization of the plakin domain.
Sonnenberg, A., Rojas, A.M., de Pereda, J.M.(2007) J Mol Biol 368: 1379-1391
- PubMed: 17397861
- DOI: https://doi.org/10.1016/j.jmb.2007.02.090
- Primary Citation of Related Structures:
2ODU, 2ODV - PubMed Abstract:
Plectin is a large and versatile cytoskeletal linker and member of the plakin protein family. Plakins share a conserved region called the plakin domain located near their N terminus. We have determined the crystal structure of an N-terminal fragment of the plakin domain of plectin to 2.05 A resolution. This region is adjacent to the actin-binding domain and is required for efficient binding to the integrin alpha6beta4 in hemidesmosomes. The structure is formed by two spectrin repeats connected by an alpha-helix that spans these two repeats. While the first repeat is very similar to other known structures, the second repeat is structurally different with a hydrophobic core, narrower than that in canonical spectrin repeats. Sequence analysis of the plakin domain revealed the presence of up to nine consecutive spectrin repeats organized in an array of tandem modules, and a Src-homology 3 domain inserted in the central spectrin repeat. The structure of the plakin domain is reminiscent of the modular organization of members of the spectrin family. The architecture of the plakin domain suggests that it forms an elongated and flexible structure, and provides a novel molecular explanation for the contribution of plectin and other plakins to the elasticity and stability of tissues subjected to mechanical stress, such as the skin and striated muscle.
Organizational Affiliation:
Netherlands Cancer Institute, Plesmanlaan 121, 1066 CX Amsterdam, The Netherlands.