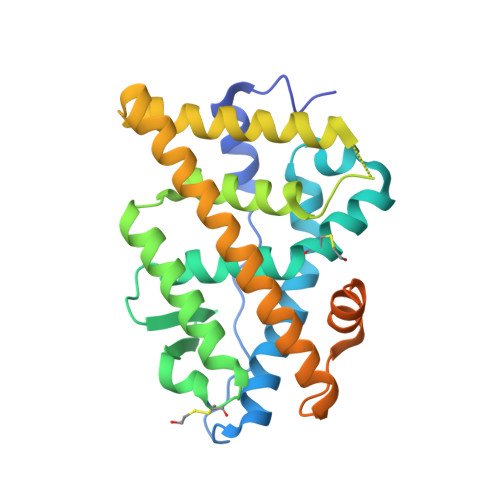
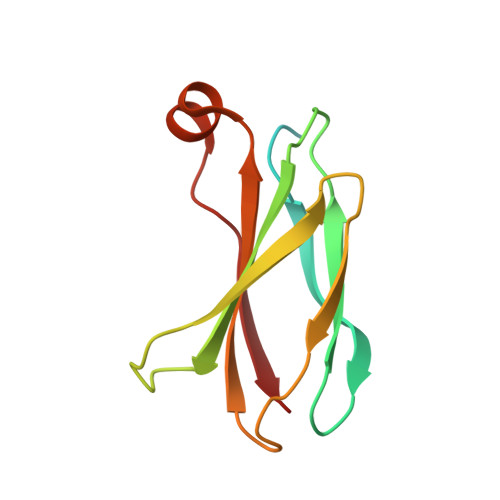
Probing protein conformational changes in living cells by using designer binding proteins: application to the estrogen receptor.
Koide, A., Abbatiello, S., Rothgery, L., Koide, S.(2002) Proc Natl Acad Sci U S A 99: 1253-1258
- PubMed: 11818562
- DOI: https://doi.org/10.1073/pnas.032665299
- Primary Citation of Related Structures:
2OCF - PubMed Abstract:
A challenge in understanding the mechanism of protein function in biology is to establish the correlation between functional form in the intracellular environment and high-resolution structures obtained with in vitro techniques. Here we present a strategy to probe conformational changes of proteins inside cells. Our method involves: (i) engineering binding proteins to different conformations of a target protein, and (ii) using them to sense changes in the surface property of the target in cells. We probed ligand-induced conformational changes of the estrogen receptor alpha (ER alpha) ligand-binding domain (LBD). By using yeast two-hybrid techniques, we first performed combinatorial library screening of "monobodies" (small antibody mimics using the scaffold of a fibronectin type III domain) for clones that bind to ER alpha and then characterized their interactions with ER alpha in the nucleus, the native environment of ER alpha, in the presence of various ligands. A library using a highly flexible loop yielded monobodies that specifically recognize a particular ligand complex of ER alpha, and the pattern of monobody specificity was consistent with the structural differences found in known crystal structures of ER alpha-LBD. A more restrained loop library yielded clones that bind both agonist- and antagonist-bound ER alpha. Furthermore, we found that a deletion of the ER alpha F domain that is C-terminally adjacent to the LBD increased the crossreactivity of monobodies to the apo-ER alpha-LBD, suggesting a dynamic nature of the ER alpha-LBD conformation and a role of the F domain in restraining the LBD in an inactive conformation.
Organizational Affiliation:
Department of Biochemistry and Biophysics, University of Rochester School of Medicine and Dentistry, 601 Elmwood Avenue, Rochester, NY 14642, USA.